A logical model provides insights into T cell receptor signaling
- PMID: 17722974
- PMCID: PMC1950951
- DOI: 10.1371/journal.pcbi.0030163
A logical model provides insights into T cell receptor signaling
Abstract
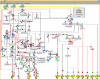
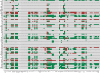
Cellular decisions are determined by complex molecular interaction networks. Large-scale signaling networks are currently being reconstructed, but the kinetic parameters and quantitative data that would allow for dynamic modeling are still scarce. Therefore, computational studies based upon the structure of these networks are of great interest. Here, a methodology relying on a logical formalism is applied to the functional analysis of the complex signaling network governing the activation of T cells via the T cell receptor, the CD4/CD8 co-receptors, and the accessory signaling receptor CD28. Our large-scale Boolean model, which comprises 94 nodes and 123 interactions and is based upon well-established qualitative knowledge from primary T cells, reveals important structural features (e.g., feedback loops and network-wide dependencies) and recapitulates the global behavior of this network for an array of published data on T cell activation in wild-type and knock-out conditions. More importantly, the model predicted unexpected signaling events after antibody-mediated perturbation of CD28 and after genetic knockout of the kinase Fyn that were subsequently experimentally validated. Finally, we show that the logical model reveals key elements and potential failure modes in network functioning and provides candidates for missing links. In summary, our large-scale logical model for T cell activation proved to be a promising in silico tool, and it inspires immunologists to ask new questions. We think that it holds valuable potential in foreseeing the effects of drugs and network modifications.
Conflict of interest statement
Figures



Similar articles
-
Elucidation of T cell signalling models.J Theor Biol. 2010 Feb 7;262(3):452-70. doi: 10.1016/j.jtbi.2009.10.017. Epub 2009 Oct 13. J Theor Biol. 2010. PMID: 19833137
-
A logical analysis of the process of T cell activation: different consequences depending on the state of CD28 engagement.J Theor Biol. 2004 Feb 21;226(4):455-66. doi: 10.1016/j.jtbi.2003.10.004. J Theor Biol. 2004. PMID: 14759651
-
A methodology for the structural and functional analysis of signaling and regulatory networks.BMC Bioinformatics. 2006 Feb 7;7:56. doi: 10.1186/1471-2105-7-56. BMC Bioinformatics. 2006. PMID: 16464248 Free PMC article.
-
Dynamic regulation of T-cell costimulation through TCR-CD28 microclusters.Immunol Rev. 2009 May;229(1):27-40. doi: 10.1111/j.1600-065X.2009.00779.x. Immunol Rev. 2009. PMID: 19426213 Review.
-
Linking molecular and cellular events in T-cell activation and synapse formation.Semin Immunol. 2003 Dec;15(6):307-15. doi: 10.1016/j.smim.2003.09.002. Semin Immunol. 2003. PMID: 15001169 Review.
Cited by
-
An Extended, Boolean Model of the Septation Initiation Network in S.Pombe Provides Insights into Its Regulation.PLoS One. 2015 Aug 5;10(8):e0134214. doi: 10.1371/journal.pone.0134214. eCollection 2015. PLoS One. 2015. PMID: 26244885 Free PMC article.
-
Microbiome modeling: a beginner's guide.Front Microbiol. 2024 Jun 19;15:1368377. doi: 10.3389/fmicb.2024.1368377. eCollection 2024. Front Microbiol. 2024. PMID: 38962127 Free PMC article. Review.
-
First insight into the kinome of human regulatory T cells.PLoS One. 2012;7(7):e40896. doi: 10.1371/journal.pone.0040896. Epub 2012 Jul 16. PLoS One. 2012. PMID: 22815858 Free PMC article.
-
A mathematical model of combined CD8 T cell costimulation by 4-1BB (CD137) and OX40 (CD134) receptors.Sci Rep. 2019 Jul 26;9(1):10862. doi: 10.1038/s41598-019-47333-y. Sci Rep. 2019. PMID: 31350431 Free PMC article.
-
Metabolic alterations impair differentiation and effector functions of CD8+ T cells.Front Immunol. 2022 Aug 2;13:945980. doi: 10.3389/fimmu.2022.945980. eCollection 2022. Front Immunol. 2022. PMID: 35983057 Free PMC article.
References
-
- Kitano H. Computational systems biology. Nature. 2002;420:206–210. - PubMed
-
- Alon U, Surette MG, Barkai N, Leibler S. Robustness in bacterial chemotaxis. Nature. 1999;397:168–171. - PubMed
-
- Wiley HS, Shvartsman SY, Lauffenburger DA. Computational modeling of the EGF-receptor system: A paradigm for systems biology. Trends Cell Biol. 2003;13:43–50. - PubMed
-
- Sasagawa S, Ozaki Y, Fujita K, Kuroda S. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat Cell Biol. 2005;7:365–373. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

