Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria
- PMID: 17722982
- PMCID: PMC1950954
- DOI: 10.1371/journal.pgen.0030138
Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria
Abstract
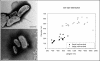
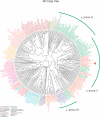
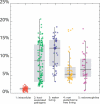
Filtration usually eliminates water-living bacteria. Here, we report on the complete genome sequence of Minibacterium massiliensis, a beta-proteobacteria that was recovered from 0.22-mum filtered water used for patients in the hospital. The unexpectedly large 4,110,251-nucleotide genome sequence of M. massiliensis was determined using the traditional shotgun sequencing approach. Bioinformatic analyses shows that the M. massiliensis genome sequence illustrates characteristic features of water-living bacteria, including overrepresentation of genes encoding transporters and transcription regulators. Phylogenomic analysis based on the gene content of available bacterial genome sequences displays a congruent evolution of water-living bacteria from various taxonomic origins, principally for genes involved in energy production and conversion, cell division, chromosome partitioning, and lipid metabolism. This phylogenomic clustering partially results from lateral gene transfer, which appears to be more frequent in water than in other environments. The M. massiliensis genome analyses strongly suggest that water-living bacteria are a common source for genes involved in heavy-metal resistance, antibiotics resistance, and virulence factors.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures


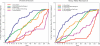
References
-
- Poindexter JS. Oligotrophy - fast and famine existence. Adv Microb Ecol. 1981;5:63–89.
-
- Iizuka T, Yamanaka S, Nishiyama T, Hiraishi A. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J Gen Appl Microbiol. 1998;44:75–84. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

