Linkage disequilibrium in wild mice
- PMID: 17722986
- PMCID: PMC1950958
- DOI: 10.1371/journal.pgen.0030144
Linkage disequilibrium in wild mice
Abstract
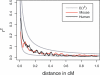
Crosses between laboratory strains of mice provide a powerful way of detecting quantitative trait loci for complex traits related to human disease. Hundreds of these loci have been detected, but only a small number of the underlying causative genes have been identified. The main difficulty is the extensive linkage disequilibrium (LD) in intercross progeny and the slow process of fine-scale mapping by traditional methods. Recently, new approaches have been introduced, such as association studies with inbred lines and multigenerational crosses. These approaches are very useful for interval reduction, but generally do not provide single-gene resolution because of strong LD extending over one to several megabases. Here, we investigate the genetic structure of a natural population of mice in Arizona to determine its suitability for fine-scale LD mapping and association studies. There are three main findings: (1) Arizona mice have a high level of genetic variation, which includes a large fraction of the sequence variation present in classical strains of laboratory mice; (2) they show clear evidence of local inbreeding but appear to lack stable population structure across the study area; and (3) LD decays with distance at a rate similar to human populations, which is considerably more rapid than in laboratory populations of mice. Strong associations in Arizona mice are limited primarily to markers less than 100 kb apart, which provides the possibility of fine-scale association mapping at the level of one or a few genes. Although other considerations, such as sample size requirements and marker discovery, are serious issues in the implementation of association studies, the genetic variation and LD results indicate that wild mice could provide a useful tool for identifying genes that cause variation in complex traits.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



Similar articles
-
Extensive linkage disequilibrium in a wild bird population.Heredity (Edinb). 2010 Jun;104(6):600-10. doi: 10.1038/hdy.2009.150. Epub 2009 Nov 4. Heredity (Edinb). 2010. PMID: 19888292
-
Levels of linkage disequilibrium in a wild bird population.Biol Lett. 2006 Sep 22;2(3):435-8. doi: 10.1098/rsbl.2006.0507. Biol Lett. 2006. PMID: 17148424 Free PMC article.
-
On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
-
The extent of linkage disequilibrium in Arabidopsis thaliana.Nat Genet. 2002 Feb;30(2):190-3. doi: 10.1038/ng813. Epub 2002 Jan 7. Nat Genet. 2002. PMID: 11780140
-
QTL mapping using high-throughput sequencing.Methods Mol Biol. 2015;1284:257-85. doi: 10.1007/978-1-4939-2444-8_13. Methods Mol Biol. 2015. PMID: 25757777 Review.
Cited by
-
Genome-wide association mapping of quantitative traits in outbred mice.G3 (Bethesda). 2012 Feb;2(2):167-74. doi: 10.1534/g3.111.001792. Epub 2012 Feb 1. G3 (Bethesda). 2012. PMID: 22384395 Free PMC article.
-
The genomic basis of environmental adaptation in house mice.PLoS Genet. 2018 Sep 24;14(9):e1007672. doi: 10.1371/journal.pgen.1007672. eCollection 2018 Sep. PLoS Genet. 2018. PMID: 30248095 Free PMC article.
-
Out of the bottleneck: the Diversity Outcross and Collaborative Cross mouse populations in behavioral genetics research.Mamm Genome. 2014 Feb;25(1-2):3-11. doi: 10.1007/s00335-013-9492-9. Epub 2013 Nov 23. Mamm Genome. 2014. PMID: 24272351 Free PMC article. Review.
-
Rapid fixation of non-native alleles revealed by genome-wide SNP analysis of hybrid tiger salamanders.BMC Evol Biol. 2009 Jul 24;9:176. doi: 10.1186/1471-2148-9-176. BMC Evol Biol. 2009. PMID: 19630983 Free PMC article.
-
Development and evaluation of the first high-throughput SNP array for common carp (Cyprinus carpio).BMC Genomics. 2014 Apr 24;15:307. doi: 10.1186/1471-2164-15-307. BMC Genomics. 2014. PMID: 24762296 Free PMC article.
References
-
- Boursot P, Auffray J, Britton-Davidian J, Bonhomme F. The evolution of house mice. Ann Rev Ecol Syst. 1993;24:119–152.
-
- Orth A, Adama T, Bonhomme F. [Natural hybridization between two subspecies of the house mouse, Mus musculus domesticus and Mus musculus castaneus, near Lake Casitas, California] Genome. 1998;41:104–110. French. - PubMed
-
- Cucchi T, Vigne JD, Auffray JC. First occurrence of the house mouse (Mus musculus domesticus Schwarz & Schwarz, 1943) in the Western Mediterranean: A zooarchaeological revision of subfossil occurrences. Biol J Linn Soc Lond. 2005;84:429–445.
-
- Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, et al. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. - PubMed
-
- Silver LM. Mouse genetics. New York: Oxford University Press; 1995. 362
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

