The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles
- PMID: 17724085
- PMCID: PMC2169053
- DOI: 10.1128/MCB.00963-07
The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles
Abstract
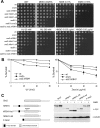
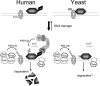
The Saccharomyces cerevisiae Srs2 UvrD DNA helicase controls genome integrity by preventing unscheduled recombination events. While Srs2 orthologues have been identified in prokaryotic and lower eukaryotic organisms, human orthologues of Srs2 have not been described so far. We found that the human F-box DNA helicase hFBH1 suppresses specific recombination defects of S. cerevisiae srs2 mutants, consistent with the finding that the helicase domain of hFBH1 is highly conserved with that of Srs2. Surprisingly, hFBH1 in the absence of SRS2 also suppresses the DNA damage sensitivity caused by inactivation of postreplication repair-dependent functions leading to PCNA ubiquitylation. The F-box domain of hFBH1, which is not present in Srs2, is crucial for hFBH1 functions in substituting for Srs2 and postreplication repair factors. Furthermore, our findings indicate that an intact F-box domain, acting as an SCF ubiquitin ligase, is required for the DNA damage-induced degradation of hFBH1 itself. Overall, our findings suggest that the hFBH1 helicase is a functional human orthologue of budding yeast Srs2 that also possesses self-regulation properties necessary to execute its recombination functions.
Figures




References
-
- Branzei, D., J. Sollier, G. Liberi, X. Zhao, D. Maeda, M. Seki, T. Enomoto, K. Ohta, and M. Foiani. 2006. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127: 509-522. - PubMed
-
- Broomfield, S., T. Hryciw, and W. Xiao. 2001. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486: 167-184. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
