Crystal structure of cefditoren complexed with Streptococcus pneumoniae penicillin-binding protein 2X: structural basis for its high antimicrobial activity
- PMID: 17724158
- PMCID: PMC2151468
- DOI: 10.1128/AAC.00743-07
Crystal structure of cefditoren complexed with Streptococcus pneumoniae penicillin-binding protein 2X: structural basis for its high antimicrobial activity
Abstract
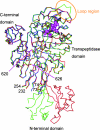
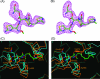
Cefditoren is the active form of cefditoren pivoxil, an oral cephalosporin antibiotic used for the treatment of respiratory tract infections and otitis media caused by bacteria such as Streptococcus pneumoniae, Haemophilus influenzae, Streptococcus pyogenes, Klebsiella pneumoniae, and methicillin-susceptible strains of Staphylococcus aureus. Beta-lactam antibiotics, including cefditoren, target penicillin-binding proteins (PBPs), which are membrane-associated enzymes that play essential roles in the peptidoglycan biosynthetic process. To envision the binding of cefditoren to PBPs, we determined the crystal structure of a trypsin-digested form of PBP 2X from S. pneumoniae strain R6 complexed with cefditoren. There are two PBP 2X molecules (designated molecules 1 and 2) per asymmetric unit. The structure reveals that the orientation of Trp374 in each molecule changes in a different way upon the formation of the complex, but each forms a hydrophobic pocket. The methylthiazole group of the C-3 side chain of cefditoren fits into this binding pocket, which consists of residues His394, Trp374, and Thr526 in molecule 1 and residues His394, Asp375, and Thr526 in molecule 2. The formation of the complex is also accompanied by an induced-fit conformational change of the enzyme in the pocket to which the C-7 side chain of cefditoren binds. These features likely play a role in the high level of activity of cefditoren against S. pneumoniae.
Figures
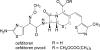

Similar articles
-
Affinity of cefditoren for penicillin-binding proteins in bacteria and its relationship with antibiotic sensitivity.Arch Microbiol. 2024 Nov 18;206(12):469. doi: 10.1007/s00203-024-04194-y. Arch Microbiol. 2024. PMID: 39556131
-
Crystal structures of biapenem and tebipenem complexed with penicillin-binding proteins 2X and 1A from Streptococcus pneumoniae.Antimicrob Agents Chemother. 2008 Jun;52(6):2053-60. doi: 10.1128/AAC.01456-07. Epub 2008 Apr 7. Antimicrob Agents Chemother. 2008. PMID: 18391040 Free PMC article.
-
Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in beta-lactam resistance in Streptococcus pneumoniae.J Mol Biol. 2006 Jan 27;355(4):684-96. doi: 10.1016/j.jmb.2005.10.030. Epub 2005 Nov 9. J Mol Biol. 2006. PMID: 16316661
-
Update of cefditoren activity tested against community-acquired pathogens associated with infections of the respiratory tract and skin and skin structures, including recent pharmacodynamic considerations.Diagn Microbiol Infect Dis. 2009 Jun;64(2):202-12. doi: 10.1016/j.diagmicrobio.2009.01.017. Epub 2009 Mar 25. Diagn Microbiol Infect Dis. 2009. PMID: 19321284 Review.
-
Cefditoren: a clinical overview.New Microbiol. 2023 Feb;46(1):9-17. New Microbiol. 2023. PMID: 36853812 Review.
Cited by
-
Cefditoren in upper and lower community-acquired respiratory tract infections.Drug Des Devel Ther. 2011 Feb 9;5:85-94. doi: 10.2147/DDDT.S9499. Drug Des Devel Ther. 2011. PMID: 21340042 Free PMC article. Review.
-
Bioactivity Studies of β-Lactam Derived Polycyclic Fused Pyrroli-Dine/Pyrrolizidine Derivatives in Dentistry: In Vitro, In Vivo and In Silico Studies.PLoS One. 2015 Jul 17;10(7):e0131433. doi: 10.1371/journal.pone.0131433. eCollection 2015. PLoS One. 2015. PMID: 26185985 Free PMC article.
-
Affinity of cefditoren for penicillin-binding proteins in bacteria and its relationship with antibiotic sensitivity.Arch Microbiol. 2024 Nov 18;206(12):469. doi: 10.1007/s00203-024-04194-y. Arch Microbiol. 2024. PMID: 39556131
-
The structures of penicillin-binding protein 4 (PBP4) and PBP5 from Enterococci provide structural insights into β-lactam resistance.J Biol Chem. 2018 Nov 30;293(48):18574-18584. doi: 10.1074/jbc.RA118.006052. Epub 2018 Oct 24. J Biol Chem. 2018. PMID: 30355734 Free PMC article.
-
Recognition of the β-lactam carboxylate triggers acylation of Neisseria gonorrhoeae penicillin-binding protein 2.J Biol Chem. 2019 Sep 20;294(38):14020-14032. doi: 10.1074/jbc.RA119.009942. Epub 2019 Jul 30. J Biol Chem. 2019. PMID: 31362987 Free PMC article.
References
-
- Barcus, V. A., K. Ghanekar, M. Yeo, T. J. Coffey, and C. G. Dowson. 1995. Genetics of high level penicillin resistance in clinical isolates of Streptococcus pneumoniae. FEMS Microbiol. Lett. 126:299-303. - PubMed
-
- Clark, C. L., K. Nagai, B. E. Dewasse, G. A. Pankuch, L. M. Ednie, M. R. Jacobs, and P. C. Appelbaum. 2002. Activity of cefditoren against respiratory pathogens. J. Antimicrob. Chemother. 50:33-41. - PubMed
-
- Contreras-Martel, C., V. Job, A. M. Di Guilmi, T. Vernet, O. Dideberg, and A. Dessen. 2006. Crystal structure of penicillin-binding protein 1a (PBP1a) reveals a mutational hotspot implicated in beta-lactam resistance in Streptococcus pneumoniae. J. Mol. Biol. 355:684-696. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

