Translational machinery of the chaetognath Spadella cephaloptera: a transcriptomic approach to the analysis of cytosolic ribosomal protein genes and their expression
- PMID: 17725830
- PMCID: PMC2020476
- DOI: 10.1186/1471-2148-7-146
Translational machinery of the chaetognath Spadella cephaloptera: a transcriptomic approach to the analysis of cytosolic ribosomal protein genes and their expression
Abstract
Background: Chaetognaths, or arrow worms, are small marine, bilaterally symmetrical metazoans. The objective of this study was to analyse ribosomal protein (RP) coding sequences from a published collection of expressed sequence tags (ESTs) from a chaetognath (Spadella cephaloptera) and to use them in phylogenetic studies.
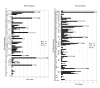
Results: This analysis has allowed us to determine the complete primary structures of 23 out of 32 RPs from the small ribosomal subunit (SSU) and 32 out of 47 RPs from the large ribosomal subunit (LSU). Ten proteins are partially determined and 14 proteins are missing. Phylogenetic analyses of concatenated RPs from six animals (chaetognath, echinoderm, mammalian, insect, mollusc and sponge) and one fungal taxa do not resolve the chaetognath phylogenetic position, although each mega-sequence comprises approximately 5,000 amino acid residues. This is probably due to the extremely biased base composition and to the high evolutionary rates in chaetognaths. However, the analysis of chaetognath RP genes revealed three unique features in the animal Kingdom. First, whereas generally in animals one RP appeared to have a single type of mRNA, two or more genes are generally transcribed for one RP type in chaetognath. Second, cDNAs with complete 5'-ends encoding a given protein sequence can be divided in two sub-groups according to a short region in their 5'-ends: two novel and highly conserved elements have been identified (5'-TAATTGAGTAGTTT-3' and 5'-TATTAAGTACTAC-3') which could correspond to different transcription factor binding sites on paralog RP genes. And, third, the overall number of deduced paralogous RPs is very high compared to those published for other animals.
Conclusion: These results suggest that in chaetognaths the deleterious effects of the presence of paralogous RPs, such as apoptosis or cancer are avoided, and also that in each protein family, some of the members could have tissue-specific and extra-ribosomal functions. These results are congruent with the hypotheses of an allopolyploid origin of this phylum and of a ribosome heterogeneity.
Figures

Similar articles
-
Careful with understudied phyla: the case of chaetognath.BMC Evol Biol. 2008 Sep 17;8:251. doi: 10.1186/1471-2148-8-251. BMC Evol Biol. 2008. PMID: 18798978 Free PMC article.
-
Selective expression of two types of 28S rRNA paralogous genes in the chaetognath Spadella cephaloptera.Cell Mol Biol (Noisy-le-grand). 2007 Sep 13;53 Suppl:OL989-93. Cell Mol Biol (Noisy-le-grand). 2007. PMID: 17877912
-
Chaetognath transcriptome reveals ancestral and unique features among bilaterians.Genome Biol. 2008;9(6):R94. doi: 10.1186/gb-2008-9-6-r94. Epub 2008 Jun 4. Genome Biol. 2008. PMID: 18533022 Free PMC article.
-
Are ribosomal proteins present at transcription sites on or off ribosomal subunits?Biochem Soc Trans. 2010 Dec;38(6):1543-7. doi: 10.1042/BST0381543. Biochem Soc Trans. 2010. PMID: 21118123 Review.
-
Lophotrochozoan relationships and parasites. A snap-shot.Parasite. 2008 Sep;15(3):329-32. doi: 10.1051/parasite/2008153329. Parasite. 2008. PMID: 18814703 Review.
Cited by
-
Translational machinery of senegalese sole (Solea senegalensis Kaup) and Atlantic halibut (Hippoglossus hippoglossus L.): comparative sequence analysis of the complete set of 60s ribosomal proteins and their expression.Mar Biotechnol (NY). 2008 Nov-Dec;10(6):676-91. doi: 10.1007/s10126-008-9104-y. Epub 2008 May 14. Mar Biotechnol (NY). 2008. PMID: 18478294
-
Careful with understudied phyla: the case of chaetognath.BMC Evol Biol. 2008 Sep 17;8:251. doi: 10.1186/1471-2148-8-251. BMC Evol Biol. 2008. PMID: 18798978 Free PMC article.
-
Transcriptome Analysis of ESTs from a Chaetognath Reveals a Deep-Branching Clade of Retrovirus-Like Retrotransposons.Open Virol J. 2008;2:44-60. doi: 10.2174/1874357900802010044. Epub 2008 May 7. Open Virol J. 2008. PMID: 19440464 Free PMC article.
-
Genome-Wide Identification, Evolution and Expression of the Complete Set of Cytoplasmic Ribosomal Protein Genes in Nile Tilapia.Int J Mol Sci. 2020 Feb 12;21(4):1230. doi: 10.3390/ijms21041230. Int J Mol Sci. 2020. PMID: 32059409 Free PMC article.
References
-
- Casanova J-P. Chaetognatha. In: Boltovskoy D, editor. South Atlantic Zooplankton. Leiden: Backhuys Publishers; 1999. pp. 1353–1374.
-
- Reeve MR. The biology of chaetognatha, I. Quantitative aspects of growth and egg production in Sagitta hispida. In: Steele JH, editor. Marine Food Chains. Edinburgh: Oliver and Boyd; 1970. pp. 168–189.
-
- Casanova J-P, Duvert M, Perez Y. Phylogenetic interest of the "chaetognath model". Mésogée. 2001;59:27–31.
-
- Casanova J-P, Duvert M, Goto T. Emergence of limb-like appendages from fins in chaetognaths. CR Acad Sci, Paris, Sci vie. 1995;318:1167–1172.
-
- Casanova J-P, Duvert M. Comparative studies and evolution of muscles in chaetognaths. Mar Biol. 2002;141:925–938. doi: 10.1007/s00227-002-0889-3. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

