Identifying genes preferentially expressed in undifferentiated embryonic stem cells
- PMID: 17725840
- PMCID: PMC1995199
- DOI: 10.1186/1471-2121-8-37
Identifying genes preferentially expressed in undifferentiated embryonic stem cells
Abstract
Background: The mechanism involved in the maintenance and differentiation of embryonic stem (ES) cells is incompletely understood.
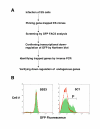
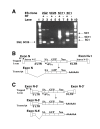
Results: To address this issue, we have developed a retroviral gene trap vector that can target genes expressed in undifferentiated ES cells. This gene trap vector harbors both GFP and Neo reporter genes. G-418 drug resistance was used to select ES clones in which the vector was integrated into transcriptionally active loci. This was then followed by GFP FACS profiling to identify ES clones with reduced GFP fluorescence and, hence, reduced transcriptional activity when ES cells differentiate. Reduced expression of the GFP reporter in six of three hundred ES clones in our pilot screening was confirmed to be down-regulated by Northern blot analysis during ES cell differentiation. These six ES clones represent four different genes. Among the six integration sites, one was at Zfp-57 whose gene product is known to be enriched in undifferentiated ES cells. Three were located in an intron of a novel isoform of CSL/RBP-J kappa which encodes the key transcription factor of the LIN-12/Notch pathway. Another was inside a gene that may encode noncoding RNA transcripts. The last integration event occurred at a locus that may harbor a novel gene.
Conclusion: Taken together, we demonstrate the use of a novel retroviral gene trap vector in identifying genes preferentially expressed in undifferentiated ES cells.
Figures



Similar articles
-
Novel vectors for homologous recombination strategies in mouse embryonic stem cells: an ES cell line expressing EGFP under control of the 5T4 promoter.Exp Cell Res. 2007 Oct 1;313(16):3604-15. doi: 10.1016/j.yexcr.2007.07.021. Epub 2007 Jul 28. Exp Cell Res. 2007. PMID: 17765223
-
Stable suppression of gene expression in murine embryonic stem cells by RNAi directed from DNA vector-based short hairpin RNA.Stem Cells. 2004;22(1):93-9. doi: 10.1634/stemcells.22-1-93. Stem Cells. 2004. PMID: 14688395
-
Transcriptional profiling of reporter genes used for molecular imaging of embryonic stem cell transplantation.Physiol Genomics. 2006 Mar 13;25(1):29-38. doi: 10.1152/physiolgenomics.00254.2005. Epub 2006 Jan 3. Physiol Genomics. 2006. PMID: 16390873
-
Mouse gene trap approach: identification of novel genes and characterization of their biological functions.Biochem Cell Biol. 1998;76(6):1029-37. Biochem Cell Biol. 1998. PMID: 10392714 Review.
-
[Genetic screening for novel genes by insertional mutagenesis with gene trap method in ES cells].Tanpakushitsu Kakusan Koso. 1995 Oct;40(14):2017-24. Tanpakushitsu Kakusan Koso. 1995. PMID: 8532857 Review. Japanese. No abstract available.
Cited by
-
Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain.J Biol Chem. 2012 Jan 13;287(3):2107-18. doi: 10.1074/jbc.M111.322644. Epub 2011 Dec 5. J Biol Chem. 2012. PMID: 22144682 Free PMC article.
-
Survival of human embryonic stem cells implanted in the guinea pig auditory epithelium.Sci Rep. 2017 Apr 7;7:46058. doi: 10.1038/srep46058. Sci Rep. 2017. PMID: 28387239 Free PMC article.
-
Maintaining memory of silencing at imprinted differentially methylated regions.Cell Mol Life Sci. 2016 May;73(9):1871-9. doi: 10.1007/s00018-016-2157-6. Epub 2016 Feb 16. Cell Mol Life Sci. 2016. PMID: 26883803 Free PMC article. Review.
-
Transient neonatal diabetes, ZFP57, and hypomethylation of multiple imprinted loci: a detailed follow-up.Diabetes Care. 2013 Mar;36(3):505-12. doi: 10.2337/dc12-0700. Epub 2012 Nov 12. Diabetes Care. 2013. PMID: 23150280 Free PMC article.
-
RBPJ, the major transcriptional effector of Notch signaling, remains associated with chromatin throughout mitosis, suggesting a role in mitotic bookmarking.PLoS Genet. 2014 Mar 6;10(3):e1004204. doi: 10.1371/journal.pgen.1004204. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603501 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

