Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent
- PMID: 17726022
- PMCID: PMC3145109
- DOI: 10.1074/jbc.M703451200
Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent
Abstract
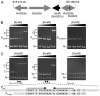
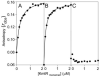
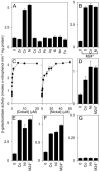
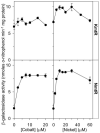
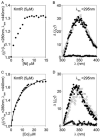
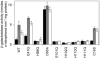
A novel ArsR-SmtB family transcriptional repressor, KmtR, has been characterized from mycobacteria. Mutants of Mycobacterium tuberculosis lacking kmtR show elevated expression of Rv2025c encoding a deduced CDF-family metal exporter. KmtR-dependent repression of the cdf and kmtR operator-promoters was alleviated by nickel and cobalt in minimal medium. Electrophoretic mobility shift assays and fluorescence anisotropy show binding of purified KmtR to nucleotide sequences containing a region of dyad symmetry from the cdf and kmtR operator-promoters. Incubation of KmtR with cobalt inhibits DNA complex assembly and metal-protein binding was confirmed. KmtR is the second, to NmtR, characterized ArsR-SmtB sensor of nickel and cobalt from M. tuberculosis suggesting special significance for these ions in this pathogen. KmtR-dependent expression is elevated in complete medium with no increase in response to metals, whereas NmtR retains a response to nickel and cobalt under these conditions. KmtR has tighter affinities for nickel and cobalt than NmtR consistent with basal levels of these metals being sensed by KmtR but not NmtR in complete medium. More than a thousand genes encoding ArsR-SmtB-related proteins are listed in databases. KmtR has none of the previously defined metal-sensing sites. Substitution of His88, Glu101, His102, His110, or His111 with Gln generated KmtR variants that repress the cdf and kmtR operator-promoters even in elevated nickel and cobalt, revealing a new sensory site. Importantly, ArsR-SmtB sequence groupings do not correspond with the different sensory motifs revealing that only the latter should be used to predict metal sensing.
Figures

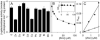



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

