siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae
- PMID: 17726046
- PMCID: PMC2094067
- DOI: 10.1093/nar/gkm646
siRNA-dependent and -independent post-transcriptional cosuppression of the LTR-retrotransposon MAGGY in the phytopathogenic fungus Magnaporthe oryzae
Abstract
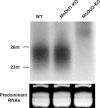
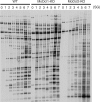
The LTR-retrotransposon MAGGY was introduced into naive genomes of Magnaporthe oryzae with different genetic backgrounds (wild-type, and MoDcl1 [mdl1] and MoDcl2 [mdl2] dicer mutants). The MoDcl2 mutants deficient in MAGGY siRNA biogenesis generally showed greater MAGGY mRNA accumulation and more rapid increase in MAGGY copy number than did the wild-type and MoDcl1 mutants exhibiting normal MAGGY siRNA accumulation, indicating that RNA silencing functioned as an effective defense against the invading element. Interestingly, however, regardless of genetic background, the rate of MAGGY transposition drastically decreased as its copy number in the genome increased. Notably, in the MoDcl2 mutant, copy-number-dependent MAGGY suppression occurred without a reduction in its mRNA accumulation, and therefore by a silencing mechanism distinct from both transcriptional gene silencing and siRNA-mediated RNA silencing. This might imply that some mechanism possibly similar to post-transcriptional cosuppression of Ty1 retrotransposition in Saccharomyces cerevisiae, which operates regardless of the abundance of target transcript and independent of RNA silencing, would also function in M. oryzae that possesses the RNA silencing machinery.
Figures
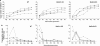


Similar articles
-
The C-terminal chromodomain-like module in the integrase domain is crucial for high transposition efficiency of the retrotransposon MAGGY.FEBS Lett. 2005 Jan 17;579(2):488-92. doi: 10.1016/j.febslet.2004.12.017. FEBS Lett. 2005. PMID: 15642364
-
Methylation is not the main force repressing the retrotransposon MAGGY in Magnaporthe grisea.Nucleic Acids Res. 2001 Mar 15;29(6):1278-84. doi: 10.1093/nar/29.6.1278. Nucleic Acids Res. 2001. PMID: 11238993 Free PMC article.
-
One of the two Dicer-like proteins in the filamentous fungi Magnaporthe oryzae genome is responsible for hairpin RNA-triggered RNA silencing and related small interfering RNA accumulation.J Biol Chem. 2004 Oct 22;279(43):44467-74. doi: 10.1074/jbc.M408259200. Epub 2004 Aug 10. J Biol Chem. 2004. PMID: 15304480
-
RNA silencing in fungi: mechanisms and applications.FEBS Lett. 2005 Oct 31;579(26):5950-7. doi: 10.1016/j.febslet.2005.08.016. Epub 2005 Aug 24. FEBS Lett. 2005. PMID: 16137680 Review.
-
The Ty1 LTR-Retrotransposon of Budding Yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0053-2014. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 26104690 Review.
Cited by
-
Copy number-dependent DNA methylation of the Pyricularia oryzae MAGGY retrotransposon is triggered by DNA damage.Commun Biol. 2021 Mar 19;4(1):351. doi: 10.1038/s42003-021-01836-5. Commun Biol. 2021. PMID: 33742058 Free PMC article.
-
The Evolutionary Significance of RNAi in the Fungal Kingdom.Int J Mol Sci. 2020 Dec 8;21(24):9348. doi: 10.3390/ijms21249348. Int J Mol Sci. 2020. PMID: 33302447 Free PMC article. Review.
-
Transcriptional control and protein specialization have roles in the functional diversification of two dicer-like proteins in Magnaporthe oryzae.Genetics. 2008 Oct;180(2):1245-9. doi: 10.1534/genetics.108.093922. Epub 2008 Sep 14. Genetics. 2008. PMID: 18791228 Free PMC article.
-
Fungal virulence and development is regulated by alternative pre-mRNA 3'end processing in Magnaporthe oryzae.PLoS Pathog. 2011 Dec;7(12):e1002441. doi: 10.1371/journal.ppat.1002441. Epub 2011 Dec 15. PLoS Pathog. 2011. PMID: 22194688 Free PMC article.
-
Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae.BMC Genomics. 2011 Jun 2;12:288. doi: 10.1186/1471-2164-12-288. BMC Genomics. 2011. PMID: 21635781 Free PMC article.
References
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
-
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. - PubMed
-
- Couch BC, Kohn LM. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea. Mycologia. 2002;94:683–693. - PubMed
-
- Kato H, Yamamoto M, Yamaguchi-Ozaki T, Kadouchi H, Iwamoto Y, Nakayashiki H, Tosa Y, Mayama S, Mori N. Pathogenicity, mating ability and DNA restriction fragment length polymorphisms of Pyricularia populations isolated from Gramineae, Bambusideae and Zingiberaceae plants. J. Gen. Plant Pathol. 2000;66:30–47.

