Efficient expression of gene variants that harbour AGA codons next to the initiation codon
- PMID: 17726048
- PMCID: PMC2034473
- DOI: 10.1093/nar/gkm643
Efficient expression of gene variants that harbour AGA codons next to the initiation codon
Abstract
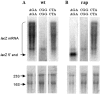
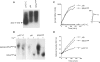
In an effort to improve the knowledge about the rules which direct the effect of the early ORF sequences on translation efficiency, we have analyzed the effect of pairs of the six arginine codons at the second and third positions on the expression of lacZ variants. Whereas the pairs of identical AGA or AGG codons were favorable for the gene expression, identical pairs of each of the four CGN codons were very inefficient. This result was unexpected because tandems of AGA or AGG codons located in more internal gene positions provoke deficient expression whilst internally located CGU and CGC are the most abundant and efficiently translated arginine codons. The mixed combinations of AGA and each of the CGN codons usually resulted in efficient rates of lacZ expression independently of the peptidyl-tRNA propensity to dissociate from the ribosome. Thus, the variant harboring the pair of AGA codons was expressed as efficiently as the variant carrying a pair of AAA codons in the same positions, a configuration reported as one of the most common and efficient for gene expression. We explain these results assuming that the presence of adenines in these early positions enhance gene expression. As expected, specific mRNA levels correlated with the intensity of lacZ expression for each variant. However, the induction of lacZ AGA AGA gene in pth cells accumulated peptidyl-tRNA(Arg4) as well as a short 5'-proximal lacZ mRNA fragment suggesting ribosome stalling due to depletion of aminoacylated-tRNA(Arg4).
Figures
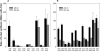
References
-
- Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 1992;6:1219–1229. - PubMed
-
- McCarthy JE, Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

