The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP
- PMID: 17726049
- PMCID: PMC2034469
- DOI: 10.1093/nar/gkm642
The p300/CBP-associated factor (PCAF) is a cofactor of ATF4 for amino acid-regulated transcription of CHOP
Abstract
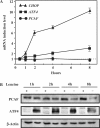
When an essential amino acid is limited, a signaling cascade is triggered that leads to increased translation of the 'master regulator', activating transcription factor 4 (ATF4), and resulting in the induction of specific target genes. Binding of ATF4 to the amino acid response element (AARE) is an essential step in the transcriptional activation of CHOP (a CCAAT/enhancer-binding protein-related gene) by amino acid deprivation. We set out to identify proteins that interact with ATF4 and that play a role in the transcriptional activation of CHOP. Using a tandem affinity purification (TAP) tag approach, we identified p300/CBP-associated factor (PCAF) as a novel interaction partner of ATF4 in leucine-starved cells. We show that the N-terminal region of ATF4 is required for a direct interaction with PCAF and demonstrate that PCAF is involved in the full transcriptional response of CHOP by amino acid starvation. Chromatin immunoprecipitation analysis revealed that PCAF is engaged on the CHOP AARE in response to amino acid starvation and that ATF4 is essential for its recruitment. We also show that PCAF stimulates ATF4-driven transcription via its histone acetyltransferase domain. Thus PCAF acts as a coactivator of ATF4 and is involved in the enhancement of CHOP transcription following amino acid starvation.
Figures





Similar articles
-
Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation.J Biol Chem. 2004 Feb 13;279(7):5288-97. doi: 10.1074/jbc.M311862200. Epub 2003 Dec 1. J Biol Chem. 2004. PMID: 14630918
-
ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation.Nucleic Acids Res. 2007;35(4):1312-21. doi: 10.1093/nar/gkm038. Epub 2007 Jan 31. Nucleic Acids Res. 2007. PMID: 17267404 Free PMC article.
-
The C/ebp-Atf response element (CARE) location reveals two distinct Atf4-dependent, elongation-mediated mechanisms for transcriptional induction of aminoacyl-tRNA synthetase genes in response to amino acid limitation.Nucleic Acids Res. 2016 Nov 16;44(20):9719-9732. doi: 10.1093/nar/gkw667. Epub 2016 Jul 28. Nucleic Acids Res. 2016. PMID: 27471030 Free PMC article.
-
ATF4-dependent transcription mediates signaling of amino acid limitation.Trends Endocrinol Metab. 2009 Nov;20(9):436-43. doi: 10.1016/j.tem.2009.05.008. Epub 2009 Sep 30. Trends Endocrinol Metab. 2009. PMID: 19800252 Free PMC article. Review.
-
Amino acid limitation regulates gene expression.Proc Nutr Soc. 1999 Aug;58(3):625-32. doi: 10.1017/s0029665199000828. Proc Nutr Soc. 1999. PMID: 10604196 Review.
Cited by
-
Neurodegeneration: Keeping ATF4 on a Tight Leash.Front Cell Neurosci. 2017 Dec 15;11:410. doi: 10.3389/fncel.2017.00410. eCollection 2017. Front Cell Neurosci. 2017. PMID: 29326555 Free PMC article. Review.
-
Is there a common upstream link for autophagic and apoptotic cell death in human high-grade gliomas?Neuro Oncol. 2011 Jul;13(7):725-35. doi: 10.1093/neuonc/nor053. Neuro Oncol. 2011. PMID: 21727211 Free PMC article.
-
Spatial Transcriptomic Study Reveals Heterogeneous Metabolic Adaptation and a Role of Pericentral PPARα/CAR/Ces2a Axis During Fasting in Mouse Liver.Adv Sci (Weinh). 2024 Nov;11(41):e2405240. doi: 10.1002/advs.202405240. Epub 2024 Sep 5. Adv Sci (Weinh). 2024. PMID: 39234807 Free PMC article.
-
Human CHAC1 Protein Degrades Glutathione, and mRNA Induction Is Regulated by the Transcription Factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element.J Biol Chem. 2015 Jun 19;290(25):15878-15891. doi: 10.1074/jbc.M114.635144. Epub 2015 Apr 30. J Biol Chem. 2015. PMID: 25931127 Free PMC article.
-
Dynamic changes in genomic histone association and modification during activation of the ASNS and ATF3 genes by amino acid limitation.Biochem J. 2013 Jan 1;449(1):219-29. doi: 10.1042/BJ20120958. Biochem J. 2013. PMID: 22978410 Free PMC article.
References
-
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. - PubMed
-
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. - PubMed
-
- Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

