Tissue-dependent paired expression of miRNAs
- PMID: 17726050
- PMCID: PMC2034466
- DOI: 10.1093/nar/gkm641
Tissue-dependent paired expression of miRNAs
Abstract
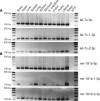
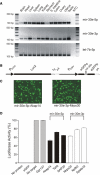
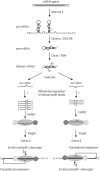
It is believed that depending on the thermodynamic stability of the 5'-strand and the 3'-strand in the stem-loop structure of a precursor microRNA (pre-miRNA), cells preferentially select the less stable one (called the miRNA or guide strand) and destroy the other one (called the miRNA* or passenger strand). However, our expression profiling analyses revealed that both strands could be co-accumulated as miRNA pairs in some tissues while being subjected to strand selection in other tissues. Our target prediction and validation assays demonstrated that both strands of a miRNA pair could target equal numbers of genes and that both were able to suppress the expression of their target genes. Our finding not only suggests that the numbers of miRNAs and their targets are much greater than what we previously thought, but also implies that novel mechanisms are involved in the tissue-dependent miRNA biogenesis and target selection process.
Figures

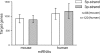
Similar articles
-
The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes.RNA. 2009 Dec;15(12):2219-35. doi: 10.1261/rna.1646909. Epub 2009 Oct 27. RNA. 2009. PMID: 19861421 Free PMC article.
-
Prediction of guide strand of microRNAs from its sequence and secondary structure.BMC Bioinformatics. 2009 Apr 9;10:105. doi: 10.1186/1471-2105-10-105. BMC Bioinformatics. 2009. PMID: 19358699 Free PMC article.
-
Profiling microRNAs and their targets in an important fleshy fruit: tomato (Solanum lycopersicum).Gene. 2014 Feb 10;535(2):198-203. doi: 10.1016/j.gene.2013.11.034. Epub 2013 Dec 4. Gene. 2014. PMID: 24315821
-
microRNA strand selection: Unwinding the rules.Wiley Interdiscip Rev RNA. 2021 May;12(3):e1627. doi: 10.1002/wrna.1627. Epub 2020 Sep 20. Wiley Interdiscip Rev RNA. 2021. PMID: 32954644 Free PMC article. Review.
-
Regulation of miRNA strand selection: follow the leader?Biochem Soc Trans. 2014 Aug;42(4):1135-40. doi: 10.1042/BST20140142. Biochem Soc Trans. 2014. PMID: 25110015 Review.
Cited by
-
MicroRNAs: Key Regulators in the Central Nervous System and Their Implication in Neurological Diseases.Int J Mol Sci. 2016 May 28;17(6):842. doi: 10.3390/ijms17060842. Int J Mol Sci. 2016. PMID: 27240359 Free PMC article. Review.
-
RNA-protein analysis using a conditional CRISPR nuclease.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):5416-21. doi: 10.1073/pnas.1302807110. Epub 2013 Mar 14. Proc Natl Acad Sci U S A. 2013. PMID: 23493562 Free PMC article.
-
MicroRNA-34 directly targets pair-rule genes and cytoskeleton component in the honey bee.Sci Rep. 2017 Jan 18;7:40884. doi: 10.1038/srep40884. Sci Rep. 2017. PMID: 28098233 Free PMC article.
-
Micromanaging vascular biology: tiny microRNAs play big band.J Vasc Res. 2009;46(6):527-40. doi: 10.1159/000226221. Epub 2009 Jun 30. J Vasc Res. 2009. PMID: 19571573 Free PMC article. Review.
-
MicroRNA‑25‑3p regulates human nucleus pulposus cell proliferation and apoptosis in intervertebral disc degeneration by targeting Bim.Mol Med Rep. 2020 Nov;22(5):3621-3628. doi: 10.3892/mmr.2020.11483. Epub 2020 Sep 2. Mol Med Rep. 2020. PMID: 32901887 Free PMC article.
References
-
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

