Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes
- PMID: 17727704
- PMCID: PMC2375005
- DOI: 10.1186/gb-2007-8-8-r175
Disruption and pseudoautosomal localization of the major histocompatibility complex in monotremes
Abstract
Background: The monotremes, represented by the duck-billed platypus and the echidnas, are the most divergent species within mammals, featuring a flamboyant mix of reptilian, mammalian and specialized characteristics. To understand the evolution of the mammalian major histocompatibility complex (MHC), the analysis of the monotreme genome is vital.
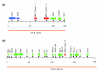
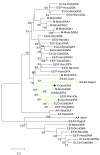
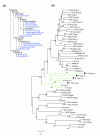
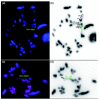
Results: We characterized several MHC containing bacterial artificial chromosome clones from platypus (Ornithorhynchus anatinus) and the short-beaked echidna (Tachyglossus aculeatus) and mapped them onto chromosomes. We discovered that the MHC of monotremes is not contiguous and locates within pseudoautosomal regions of two pairs of their sex chromosomes. The analysis revealed an MHC core region with class I and class II genes on platypus and echidna X3/Y3. Echidna X4/Y4 and platypus Y4/X5 showed synteny to the human distal class III region and beyond. We discovered an intron-containing class I pseudogene on platypus Y4/X5 at a genomic location equivalent to the human HLA-B,C region, suggesting ancestral synteny of the monotreme MHC. Analysis of male meioses from platypus and echidna showed that MHC chromosomes occupy different positions in the meiotic chains of either species.
Conclusion: Molecular and cytogenetic analyses reveal new insights into the evolution of the mammalian MHC and the multiple sex chromosome system of monotremes. In addition, our data establish the first homology link between chicken microchromosomes and the smallest chromosomes in the monotreme karyotype. Our results further suggest that segments of the monotreme MHC that now reside on separate chromosomes must once have been syntenic and that the complex sex chromosome system of monotremes is dynamic and still evolving.
Figures


Similar articles
-
The major histocompatibility complex in monotremes: an analysis of the evolution of Mhc class I genes across all three mammalian subclasses.Immunogenetics. 2002 Sep;54(6):381-93. doi: 10.1007/s00251-002-0484-2. Epub 2002 Jul 16. Immunogenetics. 2002. PMID: 12242589
-
Evolution of the major histocompatibility complex: Isolation of class II beta cDNAs from two monotremes, the platypus and the short-beaked echidna.Immunogenetics. 2003 Sep;55(6):402-11. doi: 10.1007/s00251-003-0598-1. Epub 2003 Aug 26. Immunogenetics. 2003. PMID: 12942212
-
Immunoglobulin genetics of Ornithorhynchus anatinus (platypus) and Tachyglossus aculeatus (short-beaked echidna).Comp Biochem Physiol A Mol Integr Physiol. 2003 Dec;136(4):811-9. doi: 10.1016/s1095-6433(03)00165-x. Comp Biochem Physiol A Mol Integr Physiol. 2003. PMID: 14667846 Review.
-
Core-SINE blocks comprise a large fraction of monotreme genomes; implications for vertebrate chromosome evolution.Chromosome Res. 2007;15(8):975-84. doi: 10.1007/s10577-007-1187-1. Epub 2008 Jan 9. Chromosome Res. 2007. PMID: 18185983
-
Tracing monotreme venom evolution in the genomics era.Toxins (Basel). 2014 Apr 2;6(4):1260-73. doi: 10.3390/toxins6041260. Toxins (Basel). 2014. PMID: 24699339 Free PMC article. Review.
Cited by
-
Insights into the evolution of mammalian telomerase: platypus TERT shares similarities with genes of birds and other reptiles and localizes on sex chromosomes.BMC Genomics. 2012 Jun 1;13:216. doi: 10.1186/1471-2164-13-216. BMC Genomics. 2012. PMID: 22655747 Free PMC article.
-
Sex chromosome evolution in amniotes: applications for bacterial artificial chromosome libraries.J Biomed Biotechnol. 2011;2011:132975. doi: 10.1155/2011/132975. Epub 2010 Oct 12. J Biomed Biotechnol. 2011. PMID: 20981143 Free PMC article. Review.
-
The Marine Mammal Class II Major Histocompatibility Complex Organization.Front Immunol. 2019 Apr 4;10:696. doi: 10.3389/fimmu.2019.00696. eCollection 2019. Front Immunol. 2019. PMID: 31019512 Free PMC article.
-
Identification of mediator complex 26 (Crsp7) gametologs on platypus X1 and Y5 sex chromosomes: a candidate testis-determining gene in monotremes?Chromosome Res. 2012 Jan;20(1):127-38. doi: 10.1007/s10577-011-9270-z. Chromosome Res. 2012. PMID: 22215486
-
Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes.Genome Res. 2008 Jun;18(6):965-73. doi: 10.1101/gr.7101908. Epub 2008 May 7. Genome Res. 2008. PMID: 18463302 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

