Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis
- PMID: 17727910
- PMCID: PMC2276668
- DOI: 10.1016/j.virol.2007.07.023
Stable cell lines expressing high levels of the herpes simplex virus type 1 LAT are refractory to caspase 3 activation and DNA laddering following cold shock induced apoptosis
Abstract
The herpes simplex virus type 1 (HSV-1) latency associated transcript (LAT) gene's anti-apoptosis activity plays a central, but not fully elucidated, role in enhancing the virus's reactivation phenotype. In transient transfection experiments, LAT increases cell survival following an apoptotic insult in the absence of other HSV-1 genes. However, the high background of untransfected cells has made it difficult to demonstrate that LAT inhibits specific apoptotic factors such as caspases. Here we report that, in mouse neuroblastoma cell lines (C1300) stably expressing high levels of LAT, cold shock induced apoptosis was blocked as judged by increased survival, protection against DNA fragmentation (by DNA ladder assay), and inhibition of caspase 3 cleavage and activation (Western blots). To our knowledge, this is the first report providing direct evidence that LAT blocks two biochemical hallmarks of apoptosis, caspase 3 cleavage and DNA laddering, in the absence of other HSV-1 gene products.
Figures

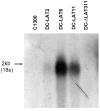
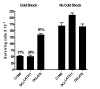
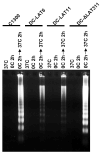

References
-
- Atanasiu D, Kent JR, Gartner JJ, Fraser NW. The stable 2-kb LAT intron of herpes simplex stimulates the expression of heat shock proteins and protects cells from stress. Virology. 2006;350(1):26–33. - PubMed
-
- Block TM, Deshmane S, Masonis J, Maggioncalda J, Valyi-Nagi T, Fraser NW. An HSV LAT null mutant reactivates slowly from latent infection and makes small plaques on CV-1 monolayers. Virology. 1993;192(2):618–630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

