Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus
- PMID: 17728234
- PMCID: PMC2168779
- DOI: 10.1128/JVI.00969-07
Nuclear magnetic resonance structure of the N-terminal domain of nonstructural protein 3 from the severe acute respiratory syndrome coronavirus
Abstract
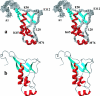
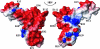
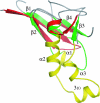
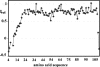
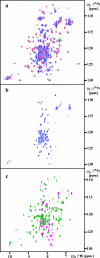
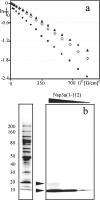
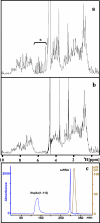
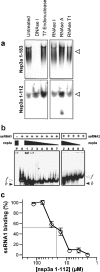
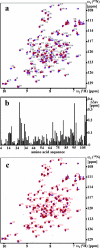
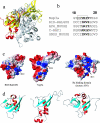
This paper describes the structure determination of nsp3a, the N-terminal domain of the severe acute respiratory syndrome coronavirus (SARS-CoV) nonstructural protein 3. nsp3a exhibits a ubiquitin-like globular fold of residues 1 to 112 and a flexibly extended glutamic acid-rich domain of residues 113 to 183. In addition to the four beta-strands and two alpha-helices that are common to ubiquitin-like folds, the globular domain of nsp3a contains two short helices representing a feature that has not previously been observed in these proteins. Nuclear magnetic resonance chemical shift perturbations showed that these unique structural elements are involved in interactions with single-stranded RNA. Structural similarities with proteins involved in various cell-signaling pathways indicate possible roles of nsp3a in viral infection and persistence.
Figures



References
-
- Altieri, A. S., D. P. Hinton, and R. A. Byrd. 1995. Association of biomolecular systems via pulsed field gradient NMR self-diffusion measurements. J. Am. Chem. Soc. 117:7566-7567.
-
- Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, J. Merz, K. M., D. M. Ferguson, D. C. Spellmyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117:5179-5197.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

