Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients
- PMID: 17728237
- PMCID: PMC2168980
- DOI: 10.1128/JVI.01059-07
Characterization of hepatitis C virus deletion mutants circulating in chronically infected patients
Abstract
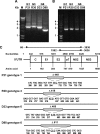
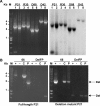
Hepatitis C virus (HCV) has a linear positive-stranded RNA genome of approximately 9,600 nucleotides in length and displays a high level of sequence diversity caused by high mutation rates and recombination. However, when we performed long distance reverse transcription-PCRs on HCV RNA isolated from serum of chronic HCV patients, not only full-length HCV genomes but also HCV RNAs which varied in size from 7,600 to 8,346 nucleotides and contained large in-frame deletions between E1 and NS2 were amplified. Carefully designed control experiments indicated that these deletion mutants are a bona fide natural RNA species, most likely packaged in virions. Moreover, deletion mutants were detected in sera of patients infected with different HCV genotypes. We observed that 7/37 (18.9%) of genotype 1, 5/43 (11.6%) of genotype 3, and 4/13 (30.7%) of genotype 6 samples contained HCV deletion mutant genomes. These observations further exemplify HCV's huge genetic diversity and warrant studies to explore their biological relevance.
Figures




Similar articles
-
Identification of novel HCV subgenome replicating persistently in chronic active hepatitis C patients.J Med Virol. 2005 Nov;77(3):399-413. doi: 10.1002/jmv.20469. J Med Virol. 2005. PMID: 16173026
-
Comparison of envelope 2 CD81 binding regions in PBMC-derived versus serum-derived hepatitis C virus isolates: higher conservation of CD81 region 2 in PBMC isolates.J Viral Hepat. 2011 Mar;18(3):181-92. doi: 10.1111/j.1365-2893.2010.01296.x. J Viral Hepat. 2011. PMID: 20367804
-
[Clinical studies on the detection of hepatitis C virus genome in the serum and the liver].Hokkaido Igaku Zasshi. 1994 Nov;69(6):1382-98. Hokkaido Igaku Zasshi. 1994. PMID: 7535730 Japanese.
-
Genetic diversity and evolution of hepatitis C virus in the Latin American region.J Clin Virol. 2005 Dec;34 Suppl 2:S1-7. doi: 10.1016/s1386-6532(05)00393-8. J Clin Virol. 2005. PMID: 16461234 Review.
-
Genetic variation of the hepatitis C virus (HCV) genome: random events or a clinically relevant issue?J Hepatol. 1993 Mar;17(3):265-8. doi: 10.1016/s0168-8278(05)80203-x. J Hepatol. 1993. PMID: 8391037 Review. No abstract available.
Cited by
-
The First Complete Genome Sequences of Hepatitis C Virus Subtype 2b from Latin America: Molecular Characterization and Phylogeographic Analysis.Viruses. 2019 Oct 31;11(11):1000. doi: 10.3390/v11111000. Viruses. 2019. PMID: 31683566 Free PMC article.
-
Are trans-complementation systems suitable for hepatitis C virus life cycle studies?J Viral Hepat. 2013 Apr;20(4):225-33. doi: 10.1111/jvh.12069. Epub 2013 Feb 14. J Viral Hepat. 2013. PMID: 23490366 Free PMC article. Review.
-
Analysis of the Enzymatic Activity of an NS3 Helicase Genotype 3a Variant Sequence Obtained from a Relapse Patient.PLoS One. 2015 Dec 10;10(12):e0144638. doi: 10.1371/journal.pone.0144638. eCollection 2015. PLoS One. 2015. PMID: 26658750 Free PMC article.
-
Generation of Defective Interfering Particles of Morbilliviruses Using Reverse Genetics.Methods Mol Biol. 2024;2808:57-70. doi: 10.1007/978-1-0716-3870-5_5. Methods Mol Biol. 2024. PMID: 38743362
-
HCV Defective Genomes Promote Persistent Infection by Modulating the Viral Life Cycle.Front Microbiol. 2018 Dec 3;9:2942. doi: 10.3389/fmicb.2018.02942. eCollection 2018. Front Microbiol. 2018. PMID: 30559733 Free PMC article.
References
-
- Aaskov, J., K. Buzacott, H. M. Thu, K. Lowry, and E. C. Holmes. 2006. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311:236-238. - PubMed
-
- Cann, A. J. (ed.). 2000. RNA viruses: a practical approach. Oxford University Press, Oxford, England.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

