Loop modeling: Sampling, filtering, and scoring
- PMID: 17729286
- PMCID: PMC2553011
- DOI: 10.1002/prot.21612
Loop modeling: Sampling, filtering, and scoring
Abstract
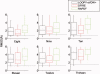
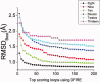
We describe a fast and accurate protocol, LoopBuilder, for the prediction of loop conformations in proteins. The procedure includes extensive sampling of backbone conformations, side chain addition, the use of a statistical potential to select a subset of these conformations, and, finally, an energy minimization and ranking with an all-atom force field. We find that the Direct Tweak algorithm used in the previously developed LOOPY program is successful in generating an ensemble of conformations that on average are closer to the native conformation than those generated by other methods. An important feature of Direct Tweak is that it checks for interactions between the loop and the rest of the protein during the loop closure process. DFIRE is found to be a particularly effective statistical potential that can bias conformation space toward conformations that are close to the native structure. Its application as a filter prior to a full molecular mechanics energy minimization both improves prediction accuracy and offers a significant savings in computer time. Final scoring is based on the OPLS/SBG-NP force field implemented in the PLOP program. The approach is also shown to be quite successful in predicting loop conformations for cases where the native side chain conformations are assumed to be unknown, suggesting that it will prove effective in real homology modeling applications.
(c) 2007 Wiley-Liss, Inc.
Figures
References
-
- Petrey D, Honig B. Protein structure prediction: inroads to biology. Mol Cell. 2005;20:811–819. - PubMed
-
- Wang Z, Moult J. SNPs, protein structure, and disease. Hum Mutat. 2001;17:263–270. - PubMed
-
- Fiser A. Protein structure modeling in the proteomics era. Expert Rev Proteomics. 2004;1:97–110. - PubMed
-
- Xiang BQ, Jia Z, Xiao FX, Zhou K, Liu P, Wei Q. The role of loop 7 in mediating calcineurin regulation. Protein Eng. 2004;16:795–798. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous