ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo
- PMID: 17760505
- PMCID: PMC1951781
- DOI: 10.1371/journal.pbio.0050232
ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo
Abstract
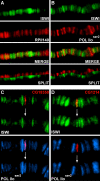
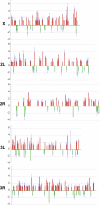
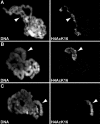
Imitation SWI (ISWI) and other ATP-dependent chromatin-remodeling factors play key roles in transcription and other processes by altering the structure and positioning of nucleosomes. Recent studies have also implicated ISWI in the regulation of higher-order chromatin structure, but its role in this process remains poorly understood. To clarify the role of ISWI in vivo, we examined defects in chromosome structure and gene expression resulting from the loss of Iswi function in Drosophila. Consistent with a broad role in transcriptional regulation, the expression of a large number of genes is altered in Iswi mutant larvae. The expression of a dominant-negative form of ISWI leads to dramatic alterations in higher-order chromatin structure, including the apparent decondensation of both mitotic and polytene chromosomes. The loss of ISWI function does not cause obvious defects in nucleosome assembly, but results in a significant reduction in the level of histone H1 associated with chromatin in vivo. These findings suggest that ISWI plays a global role in chromatin compaction in vivo by promoting the association of the linker histone H1 with chromatin.
Conflict of interest statement
Figures




References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Tremethick DJ. Higher-order structures of chromatin: The elusive 30 nm fiber. Cell. 2007;128:651–654. - PubMed
-
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding—Wrapping up transcription. Science. 2002;297:1824–1827. - PubMed
-
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. - PubMed
-
- Belmont AS. Mitotic chromosome structure and condensation. Curr Opin Cell Biol. 2006;18:632–638. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

