Programming the genome in embryonic and somatic stem cells
- PMID: 17760828
- PMCID: PMC3823245
- DOI: 10.1111/j.1582-4934.2007.00079.x
Programming the genome in embryonic and somatic stem cells
Abstract
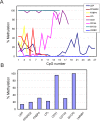
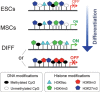
In opposition to terminally differentiated cells, stem cells can self-renew and give rise to multiple cell types. Embryonic stem cells retain the ability of the inner cell mass of blastocysts to differentiate into all cell types of the body and have acquired in culture unlimited self-renewal capacity. Somatic stem cells are found in many adult tissues, have an extensive but finite lifespan and can differentiate into a more restricted array of cell types. A growing body of evidence indicates that multi-lineage differentiation ability of stem cells can be defined by the potential for expression of lineage-specification genes. Gene expression, or as emphasized here, potential for gene expression, is largely controlled by epigenetic modifications of DNA and chromatin on genomic regulatory and coding regions. These modifications modulate chromatin organization not only on specific genes but also at the level of the whole nucleus; they can also affect timing of DNA replication. This review highlights how mechanisms by which genes are poised for transcription in undifferentiated stem cells are being uncovered through primarily the mapping of DNA methylation, histone modifications and transcription factor binding throughout the genome. The combinatorial association of epigenetic marks on developmentally regulated and lineage-specifying genes in undifferentiated cells seems to define a pluripotent state.
Figures

References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–7. - PubMed
-
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6:872–84. - PubMed
-
- Hoffman LM, Carpenter MK. Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 2005;23:699–708. - PubMed
-
- Jahagirdar BN, Verfaillie CM. Multipotent adult progenitor cell and stem cell plasticity. Stem Cell Rev. 2005;1:53–9. - PubMed
-
- Verfaillie C. Stem cell plasticity. Hematology. 2005;10:293–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

