Respiratory deficiency mediates the regulation of CHO1-encoded phosphatidylserine synthase by mRNA stability in Saccharomyces cerevisiae
- PMID: 17761681
- PMCID: PMC2150996
- DOI: 10.1074/jbc.M705098200
Respiratory deficiency mediates the regulation of CHO1-encoded phosphatidylserine synthase by mRNA stability in Saccharomyces cerevisiae
Abstract
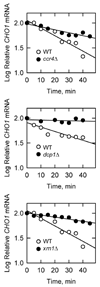
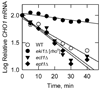
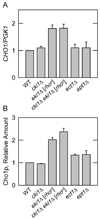
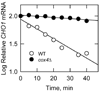
The CHO1-encoded phosphatidylserine synthase (CDP-diacylglycerol:l-serine O-phosphatidyltransferase, EC 2.7.8.8) is one of the most highly regulated phospholipid biosynthetic enzymes in the yeast Saccharomyces cerevisiae. CHO1 expression is regulated by nutrient availability through a regulatory circuit involving a UAS(INO) cis-acting element in the CHO1 promoter, the positive transcription factors Ino2p and Ino4p, and the transcriptional repressor Opi1p. In this work, we examined the post-transcriptional regulation of CHO1 by mRNA stability. CHO1 mRNA was stabilized in mutants defective in deadenylation (ccr4Delta), mRNA decapping (dcp1), and the 5'-3'-exonuclease (xrn1), indicating that the CHO1 transcript is primarily degraded through the general 5'-3' mRNA decay pathway. In respiratory-sufficient cells, the CHO1 transcript was moderately stable with a half-life of 12 min. However, the CHO1 transcript was stabilized to a half-life of >45 min in respiratory-deficient (rho(-) and rho(o)) cells, the cox4Delta mutant defective in the cytochrome c oxidase, and wild type cells treated with KCN (a cytochrome c oxidase inhibitor). The increased CHO1 mRNA stability in response to respiratory deficiency caused increases in CHO1 mRNA abundance, phosphatidylserine synthase protein and activity, and the synthesis of phosphatidylserine in vivo. Respiratory deficiency also caused increases in the activities of CDP-diacylglycerol synthase, phosphatidylserine decarboxylase, and the phospholipid methyltransferases. Phosphatidylinositol synthase and choline kinase activities were not affected by respiratory deficiency. This work advances our understanding of phosphatidylserine synthase regulation and underscores the importance of mitochondrial respiration to the regulation of phospholipid synthesis in S. cerevisiae.
Figures







Similar articles
-
Phosphatidate phosphatase regulates membrane phospholipid synthesis via phosphatidylserine synthase.Adv Biol Regul. 2018 Jan;67:49-58. doi: 10.1016/j.jbior.2017.08.001. Epub 2017 Aug 16. Adv Biol Regul. 2018. PMID: 28827025 Free PMC article. Review.
-
Regulation of phospholipid synthesis in the yeast cki1Delta eki1Delta mutant defective in the Kennedy pathway. The Cho1-encoded phosphatidylserine synthase is regulated by mRNA stability.J Biol Chem. 2004 Mar 26;279(13):12081-7. doi: 10.1074/jbc.M400297200. Epub 2004 Jan 21. J Biol Chem. 2004. PMID: 14739287
-
Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis.J Biol Chem. 2017 Aug 11;292(32):13230-13242. doi: 10.1074/jbc.M117.801720. Epub 2017 Jul 3. J Biol Chem. 2017. PMID: 28673963 Free PMC article.
-
Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc.J Biol Chem. 2004 May 21;279(21):21976-83. doi: 10.1074/jbc.M402047200. Epub 2004 Mar 17. J Biol Chem. 2004. PMID: 15028711
-
Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion.Biochim Biophys Acta. 2007 Mar;1771(3):322-30. doi: 10.1016/j.bbalip.2006.05.006. Epub 2006 May 19. Biochim Biophys Acta. 2007. PMID: 16807089 Free PMC article. Review.
Cited by
-
SUI-family genes encode phosphatidylserine synthases and regulate stem development in rice.Planta. 2013 Jan;237(1):15-27. doi: 10.1007/s00425-012-1736-5. Epub 2012 Sep 7. Planta. 2013. PMID: 22956125
-
Regulation of phospholipid synthesis in yeast.J Lipid Res. 2009 Apr;50 Suppl(Suppl):S69-73. doi: 10.1194/jlr.R800043-JLR200. Epub 2008 Oct 27. J Lipid Res. 2009. PMID: 18955729 Free PMC article. Review.
-
Phosphatidate phosphatase regulates membrane phospholipid synthesis via phosphatidylserine synthase.Adv Biol Regul. 2018 Jan;67:49-58. doi: 10.1016/j.jbior.2017.08.001. Epub 2017 Aug 16. Adv Biol Regul. 2018. PMID: 28827025 Free PMC article. Review.
-
Inferring Gene Regulatory Networks from a Population of Yeast Segregants.Sci Rep. 2019 Feb 4;9(1):1197. doi: 10.1038/s41598-018-37667-4. Sci Rep. 2019. PMID: 30718595 Free PMC article.
-
Phosphorylation of yeast phosphatidylserine synthase by protein kinase A: identification of Ser46 and Ser47 as major sites of phosphorylation.J Biol Chem. 2010 Apr 9;285(15):11526-36. doi: 10.1074/jbc.M110.100727. Epub 2010 Feb 9. J Biol Chem. 2010. PMID: 20145252 Free PMC article.
References
-
- Nikawa J, Tsukagoshi Y, Kodaki T, Yamashita S. Eur.J.Biochem. 1987;167:7–12. - PubMed
-
- Kiyono K, Miura K, Kushima Y, Hikiji T, Fukushima M, Shibuya I, Ohta A. J.Biochem. 1987;102:1089–1100. - PubMed
-
- Carman GM, Zeimetz GM. J.Biol.Chem. 1996;271:13293–13296. - PubMed
-
- Carman GM, Henry SA. Prog.Lipid Res. 1999;38:361–399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

