Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting
- PMID: 17763979
- PMCID: PMC2075529
- DOI: 10.1007/s00412-007-0118-4
Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting
Abstract
The ring-shaped cohesin complex links sister chromatids until their timely segregation during mitosis. Cohesin is enriched at centromeres where it provides the cohesive counterforce to bipolar tension produced by the mitotic spindle. As a consequence of spindle tension, centromeric sequences transiently split in pre-anaphase cells, in some organisms up to several micrometers. This 'centromere breathing' presents a paradox, how sister sequences separate where cohesin is most enriched. We now show that in the budding yeast Saccharomyces cerevisiae, cohesin binding diminishes over centromeric sequences that split during breathing. We see no evidence for cohesin translocation to surrounding sequences, suggesting that cohesin is removed from centromeres during breathing. Two pools of cohesin can be distinguished. Cohesin loaded before DNA replication, which has established sister chromatid cohesion, disappears during breathing. In contrast, cohesin loaded after DNA replication is partly retained. As sister centromeres re-associate after transient separation, cohesin is reloaded in a manner independent of the canonical cohesin loader Scc2/Scc4. Efficient centromere re-association requires the cohesion establishment factor Eco1, suggesting that re-establishment of sister chromatid cohesion contributes to the dynamic behaviour of centromeres in mitosis. These findings provide new insights into cohesin behaviour at centromeres.
Figures

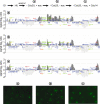
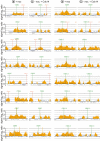


Similar articles
-
Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation.Nat Cell Biol. 2000 Aug;2(8):492-9. doi: 10.1038/35019529. Nat Cell Biol. 2000. PMID: 10934469
-
Polo kinase Cdc5 associates with centromeres to facilitate the removal of centromeric cohesin during mitosis.Mol Biol Cell. 2016 Jul 15;27(14):2286-300. doi: 10.1091/mbc.E16-01-0004. Epub 2016 May 25. Mol Biol Cell. 2016. PMID: 27226485 Free PMC article.
-
Hos1 deacetylates Smc3 to close the cohesin acetylation cycle.Mol Cell. 2010 Sep 10;39(5):677-88. doi: 10.1016/j.molcel.2010.08.009. Mol Cell. 2010. PMID: 20832720
-
Fork it over: the cohesion establishment factor Ctf7p and DNA replication.J Cell Sci. 2007 Aug 1;120(Pt 15):2471-7. doi: 10.1242/jcs.011999. J Cell Sci. 2007. PMID: 17646671 Review.
-
Shugoshin protects cohesin complexes at centromeres.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):515-21, discussion 521. doi: 10.1098/rstb.2004.1607. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897177 Free PMC article. Review.
Cited by
-
Physical Association of Saccharomyces cerevisiae Polo-like Kinase Cdc5 with Chromosomal Cohesin Facilitates DNA Damage Response.J Biol Chem. 2016 Aug 12;291(33):17228-46. doi: 10.1074/jbc.M116.727438. Epub 2016 Jun 20. J Biol Chem. 2016. PMID: 27325700 Free PMC article.
-
Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana.Chromosoma. 2009 Oct;118(5):591-605. doi: 10.1007/s00412-009-0220-x. Epub 2009 Jun 16. Chromosoma. 2009. PMID: 19533160
-
Condensin and cohesin complexity: the expanding repertoire of functions.Nat Rev Genet. 2010 Jun;11(6):391-404. doi: 10.1038/nrg2794. Epub 2010 May 5. Nat Rev Genet. 2010. PMID: 20442714 Free PMC article. Review.
-
A role for condensin in mediating transcriptional adaptation to environmental stimuli.Life Sci Alliance. 2021 Jun 3;4(7):e202000961. doi: 10.26508/lsa.202000961. Print 2021 Jul. Life Sci Alliance. 2021. PMID: 34083394 Free PMC article.
-
Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3.PLoS Genet. 2009 Sep;5(9):e1000629. doi: 10.1371/journal.pgen.1000629. Epub 2009 Sep 4. PLoS Genet. 2009. PMID: 19730685 Free PMC article.
References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1126/science.1064027', 'is_inner': False, 'url': 'https://doi.org/10.1126/science.1064027'}, {'type': 'PubMed', 'value': '11598266', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11598266/'}]}
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S0092-8674(00)81019-3', 'is_inner': False, 'url': 'https://doi.org/10.1016/s0092-8674(00)81019-3'}, {'type': 'PubMed', 'value': '10428036', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10428036/'}]}
- Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249–259 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S1097-2765(00)80420-7', 'is_inner': False, 'url': 'https://doi.org/10.1016/s1097-2765(00)80420-7'}, {'type': 'PubMed', 'value': '10882066', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10882066/'}]}
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K (2000) Cohesin’s binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5:1–20 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1038/nature02328', 'is_inner': False, 'url': 'https://doi.org/10.1038/nature02328'}, {'type': 'PubMed', 'value': '14961024', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/14961024/'}]}
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU (2004) Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 428:93–97 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1101/gad.1498707', 'is_inner': False, 'url': 'https://doi.org/10.1101/gad.1498707'}, {'type': 'PMC', 'value': 'PMC1785119', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC1785119/'}, {'type': 'PubMed', 'value': '17242156', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17242156/'}]}
- Eckert CA, Gravdahl DJ, Megee PC (2007) The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev 21:278–291 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

