Genome-wide genetic aberrations of thymoma using cDNA microarray based comparative genomic hybridization
- PMID: 17764580
- PMCID: PMC2082448
- DOI: 10.1186/1471-2164-8-305
Genome-wide genetic aberrations of thymoma using cDNA microarray based comparative genomic hybridization
Abstract
Background: Thymoma is a heterogeneous group of tumors in biology and clinical behavior. Even though thymoma is divided into five subgroups following the World Health Organization classification, the nature of the disease is mixed within the subgroups.
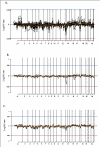
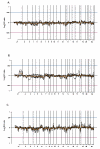
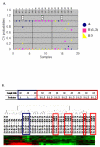
Results: We investigated the molecular characteristics of genetic changes variation of thymoma using cDNA microarray based-comparative genomic hybridization (CGH) with a 17 K cDNA microarray in an indirect, sex-matched design. Genomic DNA from the paraffin embedded 39 thymoma tissues (A 6, AB 11, B1 7, B2 7, B3 8) labeled with Cy-3 was co-hybridized with the reference placenta gDNA labeled with Cy-5. Using the CAMVS software, we investigated the deletions on chromosomes 1, 2, 3, 4, 5, 6, 8, 12, 13 and 18 throughout the thymoma. Then, we evaluated the genetic variations of thymoma based on the subgroups and the clinical behavior. First, the 36 significant genes differentiating five subgroups were selected by Significance Analysis of Microarray. Based on these genes, type AB was suggested to be heterogeneous at the molecular level as well as histologically. Next, we observed that the thymoma was divided into A, B (1, 2) and B3 subgroups with 33 significant genes. In addition, we selected 70 genes differentiating types A and B3, which differ largely in clinical behaviors. Finally, the 11 heterogeneous AB subtypes were able to correctly assign into A and B (1, 2) types based on their genetic characteristics.
Conclusion: In our study, we observed the genome-wide chromosomal aberrations of thymoma and identified significant gene sets with genetic variations related to thymoma subgroups, which might provide useful information for thymoma pathobiology.
Figures


Similar articles
-
Proteome analysis and tissue array for profiling protein markers associated with type B thymoma subclassification.Chin Med J (Engl). 2012 Aug;125(16):2811-8. Chin Med J (Engl). 2012. PMID: 22932072
-
Systematic analysis of cDNA microarray-based CGH.Int J Mol Med. 2006 Feb;17(2):261-7. Int J Mol Med. 2006. PMID: 16391824
-
High-resolution cDNA microarray CGH mapping of genomic imbalances in osteosarcoma using formalin-fixed paraffin-embedded tissue.Cytogenet Genome Res. 2004;107(1-2):77-82. doi: 10.1159/000079574. Cytogenet Genome Res. 2004. PMID: 15305059
-
High-resolution cDNA microarray-based comparative genomic hybridization analysis in neuroblastoma.Cancer Lett. 2005 Oct 18;228(1-2):71-81. doi: 10.1016/j.canlet.2004.12.056. Cancer Lett. 2005. PMID: 15951107 Review.
-
[Microarray-based comparative genomic hybridization in the study of constitutional chromosomal abnormalities].Pathol Biol (Paris). 2007 Feb;55(1):13-8. doi: 10.1016/j.patbio.2006.04.002. Epub 2006 May 11. Pathol Biol (Paris). 2007. PMID: 16697120 Review. French.
Cited by
-
The immune landscape of human thymic epithelial tumors.Nat Commun. 2022 Sep 17;13(1):5463. doi: 10.1038/s41467-022-33170-7. Nat Commun. 2022. PMID: 36115836 Free PMC article.
-
Comprehensive genomic analysis reveals clinically relevant molecular distinctions between thymic carcinomas and thymomas.Clin Cancer Res. 2009 Nov 15;15(22):6790-9. doi: 10.1158/1078-0432.CCR-09-0644. Epub 2009 Oct 27. Clin Cancer Res. 2009. PMID: 19861435 Free PMC article.
-
Single-Cell Sequencing Illuminates Thymic Development: An Updated Framework for Understanding Thymic Epithelial Tumors.Oncologist. 2024 Jun 3;29(6):473-483. doi: 10.1093/oncolo/oyae046. Oncologist. 2024. PMID: 38520743 Free PMC article. Review.
-
Artificial intelligence-powered spatial analysis of tumor-infiltrating lymphocytes as a biomarker in locally advanced unresectable thymic epithelial neoplasm: A single-center, retrospective, longitudinal cohort study.Thorac Cancer. 2023 Oct;14(30):3001-3011. doi: 10.1111/1759-7714.15089. Epub 2023 Sep 7. Thorac Cancer. 2023. PMID: 37675597 Free PMC article.
-
Thymic Epithelial Tumors phenotype relies on miR-145-5p epigenetic regulation.Mol Cancer. 2017 May 10;16(1):88. doi: 10.1186/s12943-017-0655-2. Mol Cancer. 2017. PMID: 28486946 Free PMC article.
References
-
- Bernatz PE, Harrison EG, Clagett OT. Thymoma: clinicopathologic study. J Thorac Cardiovasc Surg. 1961;42:424–44. - PubMed
-
- Kirchner T, Müller-Hermelink H-K. New approaches to the diagnosias of thymic epithelial tumors. Progr Surg Pathol. 1989;10:167–189.
-
- Suster S, Moran CA. Thymoma, atypical thymoma, and thymic carcinoma: a novel conceptual approach to the classification of thymic epithelial neoplasm. Am J Clin Pathol. 1999;111:826–833. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

