A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly
- PMID: 17765685
- PMCID: PMC2679386
- DOI: 10.1016/j.devcel.2007.07.003
A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly
Abstract
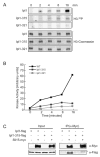
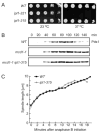
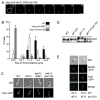
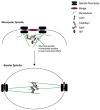
It is critical to elucidate the pathways that mediate spindle assembly and therefore ensure accurate chromosome segregation during cell division. Our studies of a unique allele of the budding yeast Ipl1/Aurora protein kinase revealed that it is required for centrosome-mediated spindle assembly in the absence of the BimC motor protein Cin8. In addition, we found that the Ase1 spindle midzone-associated protein is required for bipolar spindle assembly. The cin8 ipl1 and cin8 ase1 double mutant cells exhibit similar defects, and Ase1 overexpression completely restores spindle assembly in cin8 ipl1 strains. Consistent with the possibility that Ipl1 regulates Ase1, an ase1 mutant lacking the Ipl1 consensus phosphorylation sites cannot assemble spindles in the absence of Cin8. In addition, Ase1 phosphorylation and localization were altered in an ipl1 mutant. We therefore propose that Ipl1/Aurora and Ase1 constitute a previously unidentified spindle assembly pathway that becomes essential in the absence of Cin8.
Figures



Similar articles
-
Ipl1/Aurora-dependent phosphorylation of Sli15/INCENP regulates CPC-spindle interaction to ensure proper microtubule dynamics.J Cell Biol. 2011 Jul 11;194(1):137-53. doi: 10.1083/jcb.201009137. Epub 2011 Jul 4. J Cell Biol. 2011. PMID: 21727193 Free PMC article.
-
Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation.J Cell Biol. 2001 Nov 26;155(5):763-74. doi: 10.1083/jcb.200105029. Epub 2001 Nov 26. J Cell Biol. 2001. PMID: 11724818 Free PMC article.
-
Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae.J Cell Biol. 1999 Jun 28;145(7):1381-94. doi: 10.1083/jcb.145.7.1381. J Cell Biol. 1999. PMID: 10385519 Free PMC article.
-
Mechanisms of the Ase1/PRC1/MAP65 family in central spindle assembly.Biol Rev Camb Philos Soc. 2019 Dec;94(6):2033-2048. doi: 10.1111/brv.12547. Epub 2019 Jul 25. Biol Rev Camb Philos Soc. 2019. PMID: 31343816 Review.
-
Assembling the spindle midzone in the right place at the right time.Cell Cycle. 2008 Feb 1;7(3):283-6. doi: 10.4161/cc.7.3.5349. Epub 2007 Nov 21. Cell Cycle. 2008. PMID: 18235228 Review.
Cited by
-
Loss of function of the Cik1/Kar3 motor complex results in chromosomes with syntelic attachment that are sensed by the tension checkpoint.PLoS Genet. 2012 Feb;8(2):e1002492. doi: 10.1371/journal.pgen.1002492. Epub 2012 Feb 2. PLoS Genet. 2012. PMID: 22319456 Free PMC article.
-
Chromosome passenger complexes control anaphase duration and spindle elongation via a kinesin-5 brake.J Cell Biol. 2011 Apr 18;193(2):285-94. doi: 10.1083/jcb.201011002. Epub 2011 Apr 11. J Cell Biol. 2011. PMID: 21482719 Free PMC article.
-
The microtubule plus-end tracking protein Bik1 is required for chromosome congression.Mol Biol Cell. 2022 May 1;33(5):br7. doi: 10.1091/mbc.E21-10-0500. Epub 2022 Mar 2. Mol Biol Cell. 2022. PMID: 35235370 Free PMC article.
-
Increased Aurora B activity causes continuous disruption of kinetochore-microtubule attachments and spindle instability.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):E3996-4005. doi: 10.1073/pnas.1408017111. Epub 2014 Sep 8. Proc Natl Acad Sci U S A. 2014. PMID: 25201961 Free PMC article.
-
Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores.J Cell Sci. 2009 Dec 1;122(Pt 23):4375-82. doi: 10.1242/jcs.055566. J Cell Sci. 2009. PMID: 19923271 Free PMC article.
References
-
- Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

