Sequence-dependent DNA deformability studied using molecular dynamics simulations
- PMID: 17766249
- PMCID: PMC2094071
- DOI: 10.1093/nar/gkm627
Sequence-dependent DNA deformability studied using molecular dynamics simulations
Abstract
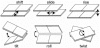
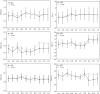
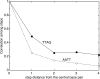
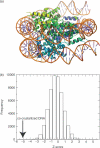
Proteins recognize specific DNA sequences not only through direct contact between amino acids and bases, but also indirectly based on the sequence-dependent conformation and deformability of the DNA (indirect readout). We used molecular dynamics simulations to analyze the sequence-dependent DNA conformations of all 136 possible tetrameric sequences sandwiched between CGCG sequences. The deformability of dimeric steps obtained by the simulations is consistent with that by the crystal structures. The simulation results further showed that the conformation and deformability of the tetramers can highly depend on the flanking base pairs. The conformations of xATx tetramers show the most rigidity and are not affected by the flanking base pairs and the xYRx show by contrast the greatest flexibility and change their conformations depending on the base pairs at both ends, suggesting tetramers with the same central dimer can show different deformabilities. These results suggest that analysis of dimeric steps alone may overlook some conformational features of DNA and provide insight into the mechanism of indirect readout during protein-DNA recognition. Moreover, the sequence dependence of DNA conformation and deformability may be used to estimate the contribution of indirect readout to the specificity of protein-DNA recognition as well as nucleosome positioning and large-scale behavior of nucleic acids.
Figures



Similar articles
-
Sequence-dependent conformational energy of DNA derived from molecular dynamics simulations: toward understanding the indirect readout mechanism in protein-DNA recognition.J Am Chem Soc. 2005 Nov 23;127(46):16074-89. doi: 10.1021/ja053241l. J Am Chem Soc. 2005. PMID: 16287294
-
Molecular dynamics simulations of B '-DNA: sequence effects on A-tract-induced bending and flexibility.J Mol Biol. 2001 Nov 16;314(1):23-40. doi: 10.1006/jmbi.2001.4926. J Mol Biol. 2001. PMID: 11724529
-
The role of DNA structure and dynamics in the recognition of bovine papillomavirus E2 protein target sequences.J Mol Biol. 2004 Jun 11;339(4):785-96. doi: 10.1016/j.jmb.2004.03.078. J Mol Biol. 2004. PMID: 15165850
-
Molecular dynamics simulations of DNA curvature and flexibility: helix phasing and premelting.Biopolymers. 2004 Feb 15;73(3):380-403. doi: 10.1002/bip.20019. Biopolymers. 2004. PMID: 14755574 Review.
-
DNA recognition and nucleosome organization.Biopolymers. 1997;44(4):423-33. doi: 10.1002/(SICI)1097-0282(1997)44:4<423::AID-BIP6>3.0.CO;2-M. Biopolymers. 1997. PMID: 9782778 Review.
Cited by
-
DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale.Nucleic Acids Res. 2013 Jul;41(Web Server issue):W56-62. doi: 10.1093/nar/gkt437. Epub 2013 May 22. Nucleic Acids Res. 2013. PMID: 23703209 Free PMC article.
-
DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences.Biophys J. 2009 Mar 18;96(6):2245-60. doi: 10.1016/j.bpj.2008.11.040. Biophys J. 2009. PMID: 19289051 Free PMC article.
-
Sequence-Dependent Fluorescence of Cy3- and Cy5-Labeled Double-Stranded DNA.Bioconjug Chem. 2016 Mar 16;27(3):840-8. doi: 10.1021/acs.bioconjchem.6b00053. Epub 2016 Feb 26. Bioconjug Chem. 2016. PMID: 26895222 Free PMC article.
-
How acidic amino acid residues facilitate DNA target site selection.Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2212501120. doi: 10.1073/pnas.2212501120. Epub 2023 Jan 12. Proc Natl Acad Sci U S A. 2023. PMID: 36634135 Free PMC article.
-
Simple Elastic Network Models for Exhaustive Analysis of Long Double-Stranded DNA Dynamics with Sequence Geometry Dependence.PLoS One. 2015 Dec 1;10(12):e0143760. doi: 10.1371/journal.pone.0143760. eCollection 2015. PLoS One. 2015. PMID: 26624614 Free PMC article.
References
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Dickerson RE. The DNA helix and how it is read. Sci. Am. 1983;249:94–111.
-
- Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. - PubMed
-
- El Hassan MA, Calladine CR. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Philos. Trans. Roy. Soc. (Ser. A) 1997;355:43–100.

