A water-explicit lattice model of heat-, cold-, and pressure-induced protein unfolding
- PMID: 17766342
- PMCID: PMC2098741
- DOI: 10.1529/biophysj.107.108530
A water-explicit lattice model of heat-, cold-, and pressure-induced protein unfolding
Abstract
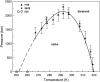
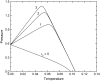
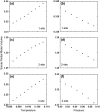
We investigate the effect of temperature and pressure on polypeptide conformational stability using a two-dimensional square lattice model in which water is represented explicitly. The model captures many aspects of water thermodynamics, including the existence of density anomalies, and we consider here the simplest representation of a protein: a hydrophobic homopolymer. We show that an explicit treatment of hydrophobic hydration is sufficient to produce cold, pressure, and thermal denaturation. We investigate the effects of the enthalpic and entropic components of the water-protein interactions on the overall folding phase diagram, and show that even a schematic model such as the one we consider yields reasonable values for the temperature and pressure ranges within which highly compact homopolymer configurations are thermodynamically stable.
Figures






Similar articles
-
Phase behavior of a lattice hydrophobic oligomer in explicit water.J Phys Chem B. 2012 Aug 9;116(31):9540-8. doi: 10.1021/jp3039237. Epub 2012 Jul 23. J Phys Chem B. 2012. PMID: 22823886
-
Role of hydrophobic hydration in protein stability: a 3D water-explicit protein model exhibiting cold and heat denaturation.J Phys Chem B. 2012 Jul 19;116(28):8095-104. doi: 10.1021/jp3039175. Epub 2012 Jul 10. J Phys Chem B. 2012. PMID: 22725973
-
The effect of sequence on the conformational stability of a model heteropolymer in explicit water.J Chem Phys. 2008 May 7;128(17):175102. doi: 10.1063/1.2909974. J Chem Phys. 2008. PMID: 18465941
-
Water mediation in protein folding and molecular recognition.Annu Rev Biophys Biomol Struct. 2006;35:389-415. doi: 10.1146/annurev.biophys.35.040405.102134. Annu Rev Biophys Biomol Struct. 2006. PMID: 16689642 Review.
-
Heat capacity in proteins.Annu Rev Phys Chem. 2005;56:521-48. doi: 10.1146/annurev.physchem.56.092503.141202. Annu Rev Phys Chem. 2005. PMID: 15796710 Review.
Cited by
-
Implicit water model within the Zimm-Bragg approach to analyze experimental data for heat and cold denaturation of proteins.Commun Chem. 2021 May 4;4(1):57. doi: 10.1038/s42004-021-00499-x. Commun Chem. 2021. PMID: 36697562 Free PMC article.
-
Hydrophobic Homopolymer's Coil-Globule Transition and Adsorption onto a Hydrophobic Surface under Different Conditions.J Phys Chem B. 2023 Jun 29;127(25):5541-5552. doi: 10.1021/acs.jpcb.3c00937. Epub 2023 Jun 19. J Phys Chem B. 2023. PMID: 37334684 Free PMC article.
-
A coarse-grained protein model in a water-like solvent.Sci Rep. 2013;3:1841. doi: 10.1038/srep01841. Sci Rep. 2013. PMID: 23674146 Free PMC article.
-
Understanding the role of hydrogen bonds in water dynamics and protein stability.J Biol Phys. 2012 Jan;38(1):27-48. doi: 10.1007/s10867-011-9235-7. Epub 2011 Oct 1. J Biol Phys. 2012. PMID: 23277668 Free PMC article.
-
Computational investigation of cold denaturation in the Trp-cage miniprotein.Proc Natl Acad Sci U S A. 2016 Aug 9;113(32):8991-6. doi: 10.1073/pnas.1607500113. Epub 2016 Jul 25. Proc Natl Acad Sci U S A. 2016. PMID: 27457961 Free PMC article.
References
-
- Hawley, S. A. 1971. Reversible pressure-temperature denaturation of chymotrypsinogen. Biochemistry. 10:2436–2442. - PubMed
-
- Heremans, K., and L. Smeller. 1998. Protein structure and dynamics at high pressure. Biochim. Biophys. Acta. 1386:353–370. - PubMed
-
- Ravindra, R., and R. Winter. 2003. On the temperature-pressure free-energy landscape of proteins. ChemPhysChem. 4:359–365. - PubMed
-
- Dill, K. A. 1990. Dominant forces in protein folding. Biochemistry. 29:7133–7155. - PubMed
-
- Kauzmann, W. 1959. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 14:1–63. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

