Crystal Structure of human pyridoxal kinase: structural basis of M(+) and M(2+) activation
- PMID: 17766369
- PMCID: PMC2204131
- DOI: 10.1110/ps.073022107
Crystal Structure of human pyridoxal kinase: structural basis of M(+) and M(2+) activation
Abstract
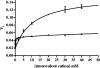
Pyridoxal kinase catalyzes the transfer of a phosphate group from ATP to the 5' alcohol of pyridoxine, pyridoxamine, and pyridoxal. In this work, kinetic studies were conducted to examine monovalent cation dependence of human pyridoxal kinase kinetic parameters. The results show that hPLK affinity for ATP and PL is increased manyfold in the presence of K(+) when compared to Na(+); however, the maximal activity of the Na(+) form of the enzyme is more than double the activity in the presence of K(+). Other monovalent cations, Li(+), Cs(+), and Rb(+) do not show significant activity. We have determined the crystal structure of hPLK in the unliganded form, and in complex with MgATP to 2.0 and 2.2 A resolution, respectively. Overall, the two structures show similar open conformation, and likely represent the catalytically idle state. The crystal structure of the MgATP complex also reveals Mg(2+) and Na(+) acting in tandem to anchor the ATP at the active site. Interestingly, the active site of hPLK acts as a sink to bind several molecules of MPD. The features of monovalent and divalent metal cation binding, active site structure, and vitamin B6 specificity are discussed in terms of the kinetic and structural studies, and are compared with those of the sheep and Escherichia coli enzymes.
Figures
Similar articles
-
Kinetic studies of the effects of K+, Na+ and Li+ on the catalytic activity of human erythrocyte pyridoxal kinase.Enzyme Protein. 1996;49(5-6):291-304. doi: 10.1159/000468639. Enzyme Protein. 1996. PMID: 9252787
-
Crystal structure of human pyridoxal kinase.J Struct Biol. 2006 Jun;154(3):327-32. doi: 10.1016/j.jsb.2006.02.008. Epub 2006 Mar 20. J Struct Biol. 2006. PMID: 16600635
-
Kinetic and structural studies of the role of the active site residue Asp235 of human pyridoxal kinase.Biochem Biophys Res Commun. 2009 Mar 27;381(1):12-5. doi: 10.1016/j.bbrc.2009.01.170. Epub 2009 Feb 4. Biochem Biophys Res Commun. 2009. PMID: 19351586
-
Vitamin B(6) salvage enzymes: mechanism, structure and regulation.Biochim Biophys Acta. 2011 Nov;1814(11):1597-608. doi: 10.1016/j.bbapap.2010.12.006. Epub 2010 Dec 20. Biochim Biophys Acta. 2011. PMID: 21182989 Review.
-
Molecular Mechanisms of Enzyme Activation by Monovalent Cations.J Biol Chem. 2016 Sep 30;291(40):20840-20848. doi: 10.1074/jbc.R116.737833. Epub 2016 Jul 26. J Biol Chem. 2016. PMID: 27462078 Free PMC article. Review.
Cited by
-
Molecular mechanisms of the non-coenzyme action of thiamin in brain: biochemical, structural and pathway analysis.Sci Rep. 2015 Jul 27;5:12583. doi: 10.1038/srep12583. Sci Rep. 2015. PMID: 26212886 Free PMC article.
-
Vitamin B₆ and Its Role in Cell Metabolism and Physiology.Cells. 2018 Jul 22;7(7):84. doi: 10.3390/cells7070084. Cells. 2018. PMID: 30037155 Free PMC article. Review.
-
Crystal structures of human pyridoxal kinase in complex with the neurotoxins, ginkgotoxin and theophylline: insights into pyridoxal kinase inhibition.PLoS One. 2012;7(7):e40954. doi: 10.1371/journal.pone.0040954. Epub 2012 Jul 18. PLoS One. 2012. PMID: 22879864 Free PMC article.
-
Chemical, genetic and structural assessment of pyridoxal kinase as a drug target in the African trypanosome.Mol Microbiol. 2012 Oct;86(1):51-64. doi: 10.1111/j.1365-2958.2012.08189.x. Epub 2012 Aug 16. Mol Microbiol. 2012. PMID: 22857512 Free PMC article.
-
Vitamin B6: a long known compound of surprising complexity.Molecules. 2009 Jan 12;14(1):329-51. doi: 10.3390/molecules14010329. Molecules. 2009. PMID: 19145213 Free PMC article. Review.
References
-
- Andersson C.E. and Mowbray, S.L. 2002. Activation of ribokinase by monovalent cations. J. Mol. Biol. 315: 409–419. - PubMed
-
- Bach S., Knockaert, M., Reinhardt, J., Lozach, O., Schmitt, S., Baratte, B., Koken, M., Coburn, S.P., Tang, L., Jiang, T., et al. 2005. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 280: 31208–31219. - PubMed
-
- Brunger A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D54: 905–921. - PubMed
-
- Campobasso N., Mathews, I.I., Begley, T.P., and Ealick, S.E. 2000. Crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry 39: 7868–7877. - PubMed
-
- Cao P., Gong, Y., Tang, L., Leung, Y.-C., and Jiang, T. 2006. Crystal structure of human pyridoxal kinase. J. Struct. Biol. 154: 327–332. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

