Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus
- PMID: 17766372
- PMCID: PMC2204132
- DOI: 10.1110/ps.072983507
Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus
Abstract
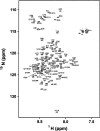
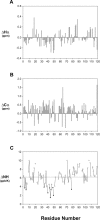
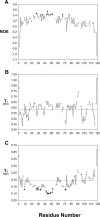
The preS1 surface antigen of hepatitis B virus (HBV) is known to play an important role in the initial attachment of HBV to hepatocytes. We have characterized structural features of the full-length preS1 using heteronuclear NMR methods and discovered that this 119-residue protein is inherently unstructured without a unique tertiary structure under a nondenaturing condition. Yet, combination of various NMR parameters shows that the preS1 contains "pre-structured" domains broadly covering its functional domains. The most prominent domain is formed by residues 27-45 and overlaps with the putative hepatocyte-binding domain (HBD) encompassing residues 21-47, within which two well-defined pre-structured motifs, formed by Pro(32)-Ala(36) and Pro(41)-Phe(45) are found. Additional, somewhat less prominent, pre-structured motifs are also formed by residues 11-18, 22-25, 37-40, and 46-50. Overall results suggest that the preS1 is a natively unstructured protein (NUP) whose N-terminal 50 residues, populated with multiple pre-structured motifs, contribute critically to hepatocyte binding.
Figures


Similar articles
-
N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus.J Hepatol. 2011 Jul;55(1):29-37. doi: 10.1016/j.jhep.2010.10.019. Epub 2010 Nov 28. J Hepatol. 2011. PMID: 21145866
-
Purification and structural analysis of the hepatitis B virus preS1 expressed from Escherichia coli.Biochem Biophys Res Commun. 2001 Apr 6;282(3):787-92. doi: 10.1006/bbrc.2001.4641. Biochem Biophys Res Commun. 2001. PMID: 11401532
-
The hepatitis B virus preS1 domain hijacks host trafficking proteins by motif mimicry.Nat Chem Biol. 2013 Sep;9(9):540-7. doi: 10.1038/nchembio.1294. Epub 2013 Jul 14. Nat Chem Biol. 2013. PMID: 23851574
-
Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes.Gastroenterology. 2005 Jul;129(1):234-45. doi: 10.1053/j.gastro.2005.03.090. Gastroenterology. 2005. PMID: 16012950
-
Attachment sites and neutralising epitopes of hepatitis B virus.Minerva Gastroenterol Dietol. 2006 Mar;52(1):3-21. Minerva Gastroenterol Dietol. 2006. PMID: 16554703 Review.
Cited by
-
The alphabet of intrinsic disorder: II. Various roles of glutamic acid in ordered and intrinsically disordered proteins.Intrinsically Disord Proteins. 2013 Apr 1;1(1):e24684. doi: 10.4161/idp.24684. eCollection 2013 Jan-Dec. Intrinsically Disord Proteins. 2013. PMID: 28516010 Free PMC article. Review.
-
Intrinsically disordered proteins in the nucleus of human cells.Biochem Biophys Rep. 2015 Mar 24;1:33-51. doi: 10.1016/j.bbrep.2015.03.003. eCollection 2015 May. Biochem Biophys Rep. 2015. PMID: 29124132 Free PMC article.
-
The transiently ordered regions in intrinsically disordered ExsE are correlated with structural elements involved in chaperone binding.Biochem Biophys Res Commun. 2012 Jan 6;417(1):129-34. doi: 10.1016/j.bbrc.2011.11.070. Epub 2011 Nov 25. Biochem Biophys Res Commun. 2012. PMID: 22138394 Free PMC article.
-
Structural basis of hepatitis B virus receptor binding.Nat Struct Mol Biol. 2024 Mar;31(3):447-454. doi: 10.1038/s41594-023-01191-5. Epub 2024 Jan 17. Nat Struct Mol Biol. 2024. PMID: 38233573
-
Choice of Force Field for Proteins Containing Structured and Intrinsically Disordered Regions.Biophys J. 2020 Apr 7;118(7):1621-1633. doi: 10.1016/j.bpj.2020.02.019. Epub 2020 Feb 29. Biophys J. 2020. PMID: 32367806 Free PMC article.
References
-
- Bax A. and Grzesiek, S. 1993. Methodological advances in protein NMR. In NMR of proteins (eds. G.M. Clore and A.M. Gronenborn), pp. 33–52. MacMillan, London, UK.
-
- Bienkiewicz E.A., Adkins, J.N., and Lumb, K.J. 2002. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1). Biochemistry 41: 752–759. - PubMed
-
- Blumberg B.S., Millman, I., Venkateswaran, P.S., and Thyagarajan, S.P. 1989. Hepatitis B virus and hepatocellular carcinoma–treatment of HBV carriers with Phyllanthus amarus. Cancer Detect. Prev. 14: 195–201. - PubMed
-
- Bochkareva E., Kaustov, L., Ayed, A., Yi, G.S., Lu, Y., Pineda-Lucena, A., Liao, J.C., Okorokov, A.L., Milner, J., Arrowsmith, C.H., et al. 2005. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. 102: 15412–15417. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

