Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation
- PMID: 17766401
- PMCID: PMC2002633
- DOI: 10.1105/tpc.107.052274
Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation
Abstract
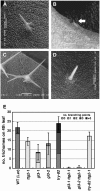
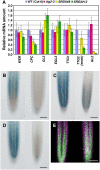
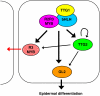
Arabidopsis thaliana TRANSPARENT TESTA GLABRA2 (TTG2) encodes a WRKY transcription factor and is expressed in young leaves, trichomes, seed coats, and root hairless cells. An examination of several trichome and root hair mutants indicates that MYB and bHLH genes regulate TTG2 expression. Two MYB binding sites in the TTG2 5' regulatory region act as cis regulatory elements and as direct targets of R2R3 MYB transcription factors such as WEREWOLF, GLABRA1, and TRANSPARENT TESTA2. Mutations in TTG2 cause phenotypic defects in trichome development and seed color pigmentation. Transgenic plants expressing a chimeric repressor version of the TTG2 protein (TTG2:SRDX) showed defects in trichome formation, anthocyanin accumulation, seed color pigmentation, and differentiation of root hairless cells. GLABRA2 (GL2) expression was markedly reduced in roots of ProTTG2:TTG2:SRDX transgenic plants, suggesting that TTG2 is involved in the regulation of GL2 expression, although GL2 expression in the ttg2 mutant was similar to that in the wild type. Our analysis suggests a new step in a regulatory cascade of epidermal differentiation, in which complexes containing R2R3 MYB and bHLH transcription factors regulate the expression of TTG2, which then regulates GL2 expression with complexes containing R2R3 MYB and bHLH in the differentiation of trichomes and root hairless cells.
Figures







References
-
- Baudry, A., Heim, M.A., Dubreucq, B., Caboche, M., Weisshaar, B., and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 366–380. - PubMed
-
- Bernhardt, C., Lee, M.M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130 6431–6439. - PubMed
-
- Czechowski, T., Bari, R.P., Stitt, M., Scheible, W.R., and Udvardi, M.K. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: Unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 38 366–379. - PubMed
-
- Di Cristina, M., Sessa, G., Dolan, L., Linstead, P., Baima, S., Ruberti, I., and Morelli, G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10 393–402. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

