Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci
- PMID: 17766420
- PMCID: PMC2168755
- DOI: 10.1128/JB.00837-07
Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci
Abstract
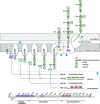
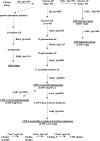
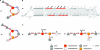
The sequences of the capsular biosynthetic (cps) loci of 90 serotypes of Streptococcus pneumoniae have recently been determined. Bioinformatic procedures were used to predict the general functions of 1,973 of the 1,999 gene products and to identify proteins within the same homology group, Pfam family, and CAZy glycosyltransferase family. Correlating cps gene content with the 54 known capsular polysaccharide (CPS) structures provided tentative assignments of the specific functions of the different homology groups of each functional class (regulatory proteins, enzymes for synthesis of CPS constituents, polymerases, flippases, initial sugar transferases, glycosyltransferases [GTs], phosphotransferases, acetyltransferases, and pyruvyltransferases). Assignment of the glycosidic linkages catalyzed by the 342 GTs (92 homology groups) is problematic, but tentative assignments could be made by using this large set of cps loci and CPS structures to correlate the presence of particular GTs with specific glycosidic linkages, by correlating inverting or retaining linkages in CPS repeat units with the inverting or retaining mechanisms of the GTs predicted from their CAZy family membership, and by comparing the CPS structures of serotypes that have very similar cps gene contents. These large-scale comparisons between structure and gene content assigned the linkages catalyzed by 72% of the GTs, and all linkages were assigned in 32 of the serotypes with known repeat unit structures. Clear examples where very similar initial sugar transferases or glycosyltransferases catalyze different linkages in different serotypes were also identified. These assignments should provide a stimulus for biochemical studies to evaluate the reactions that are proposed.
Figures


References
-
- Abbott, J. C., D. M. Aanensen, K. Rutherford, S. Butcher, and B. G. Spratt. 2005. WebACT—an online companion for the Artemis comparison tool. Bioinformatics 21:3665-3666. - PubMed
-
- Akana, J., A. A. Fedorov, E. Fedorov, W. R. Novak, P. C. Babbitt, S. C. Almo, and J. A. Gerlt. 2006. d-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ribulose-phosphate binding (beta/alpha)8-barrel superfamily. Biochemistry 45:2493-2503. - PubMed
-
- Amer, A. O., and M. A. Valvano. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571-582. - PubMed
-
- Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. López, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2000. InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16:1145-1150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

