Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci
- PMID: 17766424
- PMCID: PMC2168730
- DOI: 10.1128/JB.00836-07
Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci
Abstract
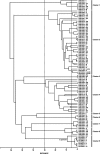
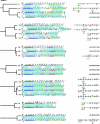
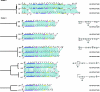
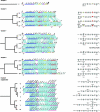
Streptococcus pneumoniae (the pneumococcus) produces 1 of 91 capsular polysaccharides (CPS) that define the serotype. The cps loci of 88 pneumococcal serotypes whose CPS is synthesized by the Wzy-dependent pathway were compared with each other and with additional streptococcal polysaccharide biosynthetic loci and were clustered according to the proportion of shared homology groups (HGs), weighted for the sequence similarities between the genes encoding the shared HGs. The cps loci of the 88 pneumococcal serotypes were distributed into eight major clusters and 21 subclusters. All serotypes within the same serogroup fell into the same major cluster, but in six cases, serotypes within the same serogroup were in different subclusters and, conversely, nine subclusters included completely different serotypes. The closely related cps loci within a subcluster were compared to the known CPS structures to relate gene content to structure. The Streptococcus oralis and Streptococcus mitis polysaccharide biosynthetic loci clustered within the pneumococcal cps loci and were in a subcluster that also included the cps locus of pneumococcal serotype 21, whereas the Streptococcus agalactiae cps loci formed a single cluster that was not closely related to any of the pneumococcal cps clusters.
Figures


References
-
- Abbott, J. C., D. M. Aanensen, K. Rutherford, S. Butcher, and B. G. Spratt. 2005. WebACT—an online companion for the Artemis comparison tool. Bioinformatics 21:3665-3666. - PubMed
-
- Batavyal, L., and N. Roy. 1983. Structure of the capsular polysaccharide of Diplococcus pneumoniae type 31. Carbohydr. Res. 119:300-302.
-
- Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

