Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil
- PMID: 17766442
- PMCID: PMC2074960
- DOI: 10.1128/AEM.01536-07
Impact of plant functional group, plant species, and sampling time on the composition of nirK-type denitrifier communities in soil
Abstract
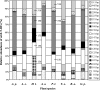
We studied the influence of eight nonleguminous grassland plant species belonging to two functional groups (grasses and forbs) on the composition of soil denitrifier communities in experimental microcosms over two consecutive years. Denitrifier community composition was analyzed by terminal restriction fragment length polymorphism (T-RFLP) of PCR-amplified nirK gene fragments coding for the copper-containing nitrite reductase. The impact of experimental factors (plant functional group, plant species, sampling time, and interactions between them) on the structure of soil denitrifier communities (i.e., T-RFLP patterns) was analyzed by canonical correspondence analysis. While the functional group of a plant did not affect nirK-type denitrifier communities, plant species identity did influence their composition. This effect changed with sampling time, indicating community changes due to seasonal conditions and a development of the plants in the microcosms. Differences in total soil nitrogen and carbon, soil pH, and root biomass were observed at the end of the experiment. However, statistical analysis revealed that the plants affected the nirK-type denitrifier community composition directly, e.g., through root exudates. Assignment of abundant T-RFs to cloned nirK sequences from the soil and subsequent phylogenetic analysis indicated a dominance of yet-unknown nirK genotypes and of genes related to nirK from denitrifiers of the order Rhizobiales. In conclusion, individual species of nonleguminous plants directly influenced the composition of denitrifier communities in soil, but environmental conditions had additional significant effects.
Figures



References
-
- Boyle, S. A., J. J. Rich, P. J. Bottomley, K. Cromack Jr., and D. D. Myrold. 2006. Reciprocal transfer effects on denitrifying community composition and activity at forest and meadow sites in the Cascade Mountains of Oregon. Soil Biol. Biochem. 38:870-878.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

