Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting
- PMID: 17767727
- PMCID: PMC1963484
- DOI: 10.1186/1471-2148-7-S2-S11
Phylogenomics of species from four genera of New World monkeys by flow sorting and reciprocal chromosome painting
Abstract
Background: The taxonomic and phylogenetic relationships of New World monkeys (Platyrrhini) are difficult to distinguish on the basis of morphology and because diagnostic fossils are rare. Recently, molecular data have led to a radical revision of the traditional taxonomy and phylogeny of these primates. Here we examine new hypotheses of platyrrhine evolutionary relationships by reciprocal chromosome painting after chromosome flow sorting of species belonging to four genera of platyrrhines included in the Cebidae family: Callithrix argentata (silvered-marmoset), Cebuella pygmaea (pygmy marmoset), Callimico goeldii (Goeldi's marmoset) and Saimiri sciureus (squirrel monkey). This is the first report of reciprocal painting in marmosets.
Results: The paints made from chromosome flow sorting of the four platyrrhine monkeys provided from 42 to 45 hybridization signals on human metaphases. The reciprocal painting of monkey probes on human chromosomes revealed that 21 breakpoints are common to all four studied species. There are only three additional breakpoints. A breakpoint on human chromosome 13 was found in Callithrix argentata, Cebuella pygmaea and Callimico goeldii, but not in Saimiri sciureus. There are two additional breakpoints on human chromosome 5: one is specific to squirrel monkeys, and the other to Goeldi's marmoset.
Conclusion: The reciprocal painting results support the molecular genomic assemblage of Cebidae. We demonstrated that the five chromosome associations previously hypothesized to phylogenetically link tamarins and marmosets are homologous and represent derived chromosome rearrangements. Four of these derived homologous associations tightly nest Callimico goeldii with marmosets. One derived association 2/15 may place squirrel monkeys within the Cebidae assemblage. An apparently common breakpoint on chromosome 5q33 found in both Saimiri and Aotus nancymae could be evidence of a phylogenetic link between these species. Comparison with previous reports shows that many syntenic associations found in platyrrhines have the same breakpoints and are homologous, derived rearrangements showing that the New World monkeys are a closely related group of species. Our data support the hypothesis that the ancestral karyotype of the Platyrrhini has a diploid number of 2n = 54 and is almost identical to that found today in capuchin monkeys; congruent with a basal position of the Cebidae among platyrrhine families.
Figures
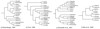





References
-
- Master JC. When, where and how? Reconstructing timeline for primate evolution using molecular and fossil data. In: Sineo L, Stanyon R, editor. Primate Cytogenetics and Comparative Genomics. Florence: Florence University Press; 2006. pp. 105–118.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

