Mutation pressure shapes codon usage in the GC-Rich genome of foot-and-mouth disease virus
- PMID: 17768673
- PMCID: PMC7089325
- DOI: 10.1007/s11262-007-0159-z
Mutation pressure shapes codon usage in the GC-Rich genome of foot-and-mouth disease virus
Abstract
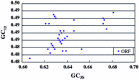
Foot-and-mouth disease (FMD) is economically the most important viral-induced livestock disease worldwide. In this study, we report the results of a survey of codon usage bias of FMD virus (FMDV) representing all seven serotypes (A, O, C, Asia 1, SAT 1, SAT 2, and SAT 3). Correspondence analysis, a commonly used multivariate statistical approach, was carried out to analyze synonymous codon usage bias. The analysis showed that the overall extent of codon usage bias in FMDV is low. Furthermore, the good correlation between the frequency of G + C at the synonymous third position of sense codons (GC3S) content at silent sites of each sequence and codon usage bias suggested that mutation pressure rather than natural (translational) selection is the most important determinant of the codon bias observed. In addition, other factors, such as the lengths of open reading frame (ORF) and the hydrophobicity of genes also influence the codon usage variation among the genomes of FMDV in a minor way. The result of phylogenetic analyses based on the relative synonymous codon usage (RSCU) values indicated a few obvious phylogenetic incongruities, which suggest that more FMDV genome diversity may exist in nature than is currently indicated. Our work might give some clues to the features of FMDV genome and some evolutionary information of this virus.
Figures
Similar articles
-
Analysis of synonymous codon usage in foot-and-mouth disease virus.Vet Res Commun. 2010 Apr;34(4):393-404. doi: 10.1007/s11259-010-9359-4. Epub 2010 Apr 28. Vet Res Commun. 2010. PMID: 20425142
-
The analysis of codon bias of foot-and-mouth disease virus and the adaptation of this virus to the hosts.Infect Genet Evol. 2013 Mar;14:105-10. doi: 10.1016/j.meegid.2012.09.020. Epub 2012 Dec 4. Infect Genet Evol. 2013. PMID: 23220329
-
An extensive analysis of Codon usage pattern, Evolutionary rate, and Phylogeographic reconstruction in Foot and mouth disease (FMD) serotypes (A, Asia 1, and O) of six major climatic zones of India: A comparative study.Acta Trop. 2022 Dec;236:106674. doi: 10.1016/j.actatropica.2022.106674. Epub 2022 Aug 30. Acta Trop. 2022. PMID: 36055369
-
Amber codon is genetically unstable in generation of premature termination codon (PTC)-harbouring Foot-and-mouth disease virus (FMDV) via genetic code expansion.RNA Biol. 2021 Dec;18(12):2330-2341. doi: 10.1080/15476286.2021.1907055. Epub 2021 Apr 14. RNA Biol. 2021. PMID: 33849391 Free PMC article.
-
The codon usage model of the context flanking each cleavage site in the polyprotein of foot-and-mouth disease virus.Infect Genet Evol. 2011 Oct;11(7):1815-9. doi: 10.1016/j.meegid.2011.07.014. Epub 2011 Jul 23. Infect Genet Evol. 2011. PMID: 21801856
Cited by
-
Analysis of codon usage in Newcastle disease virus.Virus Genes. 2011 Apr;42(2):245-53. doi: 10.1007/s11262-011-0574-z. Epub 2011 Jan 20. Virus Genes. 2011. PMID: 21249440 Free PMC article.
-
Revelation of Influencing Factors in Overall Codon Usage Bias of Equine Influenza Viruses.PLoS One. 2016 Apr 27;11(4):e0154376. doi: 10.1371/journal.pone.0154376. eCollection 2016. PLoS One. 2016. PMID: 27119730 Free PMC article.
-
Analysis of synonymous codon usage in hepatitis A virus.Virol J. 2011 Apr 16;8:174. doi: 10.1186/1743-422X-8-174. Virol J. 2011. PMID: 21496278 Free PMC article.
-
The effects of the codon usage and translation speed on protein folding of 3D(pol) of foot-and-mouth disease virus.Vet Res Commun. 2013 Sep;37(3):243-50. doi: 10.1007/s11259-013-9564-z. Epub 2013 May 29. Vet Res Commun. 2013. PMID: 23715863 No abstract available.
-
System analysis of synonymous codon usage biases in archaeal virus genomes.J Theor Biol. 2014 Aug 21;355:128-39. doi: 10.1016/j.jtbi.2014.03.022. Epub 2014 Mar 28. J Theor Biol. 2014. PMID: 24685889 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

