Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging
- PMID: 17784782
- PMCID: PMC1963511
- DOI: 10.1371/journal.pcbi.0030170
Buffering mechanisms in aging: a systems approach toward uncovering the genetic component of aging
Abstract
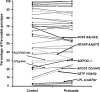
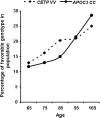
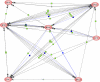
An unrealized potential to understand the genetic basis of aging in humans, is to consider the immense survival advantage of the rare individuals who live 100 years or more. The Longevity Gene Study was initiated in 1998 at the Albert Einstein College of Medicine to investigate longevity genes in a selected population: the "oldest old" Ashkenazi Jews, 95 years of age and older, and their children. The study proved the principle that some of these subjects are endowed with longevity-promoting genotypes. Here we reason that some of the favorable genotypes act as mechanisms that buffer the deleterious effect of age-related disease genes. As a result, the frequency of deleterious genotypes may increase among individuals with extreme lifespan because their protective genotype allows disease-related genes to accumulate. Thus, studies of genotypic frequencies among different age groups can elucidate the genetic determinants and pathways responsible for longevity. Borrowing from evolutionary theory, we present arguments regarding the differential survival via buffering mechanisms and their target age-related disease genes in searching for aging and longevity genes. Using more than 1,200 subjects between the sixth and eleventh decades of life (at least 140 subjects in each group), we corroborate our hypotheses experimentally. We study 66 common allelic site polymorphism in 36 candidate genes on the basis of their phenotype. Among them we have identified a candidate-buffering mechanism and its candidate age-related disease gene target. Previously, the beneficial effect of an advantageous cholesteryl ester transfer protein (CETP-VV) genotype on lipoprotein particle size in association with decreased metabolic and cardiovascular diseases, as well as with better cognitive function, have been demonstrated. We report an additional advantageous effect of the CETP-VV (favorable) genotype in neutralizing the deleterious effects of the lipoprotein(a) (LPA) gene. Finally, using literature-based interaction discovery methods, we use the set of longevity genes, buffering genes, and their age-related target disease genes to construct the underlying subnetwork of interacting genes that is expected to be responsible for longevity. Genome wide, high-throughput hypothesis-free analyses are currently being utilized to elucidate unknown genetic pathways in many model organisms, linking observed phenotypes to their underlying genetic mechanisms. The longevity phenotype and its genetic mechanisms, such as our buffering hypothesis, are similar; thus, the experimental corroboration of our hypothesis provides a proof of concept for the utility of high-throughput methods for elucidating such mechanisms. It also provides a framework for developing strategies to prevent some age-related diseases by intervention at the appropriate level.
Conflict of interest statement
Figures



References
-
- Olshansky SJ, Passaro DJ, Hershow RC, Layden J, Carnes BA, et al. A potential decline in life expectancy in the United States in the 21st century. N Engl J Med. 2005;352:1138–1145. - PubMed
-
- Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. - PubMed
-
- Larsen PL. Asking the age-old questions. Nat Genet. 2001;28:102–104. - PubMed
-
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. - PubMed
-
- Nissen H, Hansen AB, Guldberg P, Petersen NE, Larsen ML, et al. Detection of a single base deletion in codon 424 of the low density lipoprotein receptor gene in a Danish family with familial hypercholesterolemia. Atherosclerosis. 1994;111:209–215. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

