Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over
- PMID: 17784788
- PMCID: PMC1959360
- DOI: 10.1371/journal.pgen.0030139
Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over
Abstract
Crossing over during meiotic prophase I is required for sexual reproduction in mice and contributes to genome-wide genetic diversity. Here we report on the characterization of an N-ethyl-N-nitrosourea-induced, recessive allele called mei4, which causes sterility in both sexes owing to meiotic defects. In mutant spermatocytes, chromosomes fail to congress properly at the metaphase plate, leading to arrest and apoptosis before the first meiotic division. Mutant oocytes have a similar chromosomal phenotype but in vitro can undergo meiotic divisions and fertilization before arresting. During late meiotic prophase in mei4 mutant males, absence of cyclin dependent kinase 2 and mismatch repair protein association from chromosome cores is correlated with the premature separation of bivalents at diplonema owing to lack of chiasmata. We have identified the causative mutation, a transversion in the 5' splice donor site of exon 1 in the mouse ortholog of Human Enhancer of Invasion 10 (Hei10; also known as Gm288 in mouse and CCNB1IP1 in human), a putative B-type cyclin E3 ubiquitin ligase. Importantly, orthologs of Hei10 are found exclusively in deuterostomes and not in more ancestral protostomes such as yeast, worms, or flies. The cloning and characterization of the mei4 allele of Hei10 demonstrates a novel link between cell cycle regulation and mismatch repair during prophase I.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

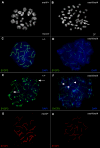
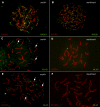
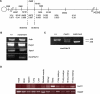

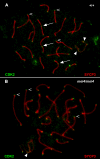

Similar articles
-
Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination.Nat Genet. 2014 Feb;46(2):194-9. doi: 10.1038/ng.2858. Epub 2014 Jan 5. Nat Genet. 2014. PMID: 24390283 Free PMC article.
-
A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels.Mol Cell Biol. 2003 Mar;23(6):2109-22. doi: 10.1128/MCB.23.6.2109-2122.2003. Mol Cell Biol. 2003. PMID: 12612082 Free PMC article.
-
MEI4 – a central player in the regulation of meiotic DNA double-strand break formation in the mouse.J Cell Sci. 2015 May 1;128(9):1800-11. doi: 10.1242/jcs.165464. Epub 2015 Mar 20. J Cell Sci. 2015. PMID: 25795304 Free PMC article.
-
A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination.Science. 2017 Jan 27;355(6323):403-407. doi: 10.1126/science.aaf6407. Epub 2017 Jan 5. Science. 2017. PMID: 28059716 Free PMC article.
-
Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals.Endocr Rev. 2006 Jun;27(4):398-426. doi: 10.1210/er.2005-0017. Epub 2006 Mar 16. Endocr Rev. 2006. PMID: 16543383 Review.
Cited by
-
The Arabidopsis HEI10 is a new ZMM protein related to Zip3.PLoS Genet. 2012;8(7):e1002799. doi: 10.1371/journal.pgen.1002799. Epub 2012 Jul 26. PLoS Genet. 2012. PMID: 22844245 Free PMC article.
-
Cis-Regulatory Evolution of CCNB1IP1 Driving Gradual Increase of Cortical Size and Folding in primates.bioRxiv [Preprint]. 2024 Dec 9:2024.12.08.627376. doi: 10.1101/2024.12.08.627376. bioRxiv. 2024. PMID: 39713381 Free PMC article. Preprint.
-
ENU mutagenesis in mice identifies candidate genes for hypogonadism.Mamm Genome. 2012 Jun;23(5-6):346-55. doi: 10.1007/s00335-011-9388-5. Epub 2012 Jan 19. Mamm Genome. 2012. PMID: 22258617 Free PMC article.
-
H2A.Z is essential for oocyte maturation and fertility in female mouse.Nat Struct Mol Biol. 2025 Jun 13. doi: 10.1038/s41594-025-01580-y. Online ahead of print. Nat Struct Mol Biol. 2025. PMID: 40514538
-
AKAP9 is essential for spermatogenesis and sertoli cell maturation in mice.Genetics. 2013 Jun;194(2):447-57. doi: 10.1534/genetics.113.150789. Epub 2013 Apr 22. Genetics. 2013. PMID: 23608191 Free PMC article.
References
-
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. - PubMed
-
- Morelli MA, Cohen PE. Not all germ cells are created equal: Aspects of sexual dimorphism in mammalian meiosis. Reproduction. 2005;130:761–781. - PubMed
-
- Keeney S, Baudat F, Angeles M, Zhou ZH, Copeland NG, et al. A mouse homolog of the Saccharomyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61:170–182. - PubMed
-
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell. 2000;6:975–987. - PubMed
-
- Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, et al. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

