Borrelia burgdorferi adhesins identified using in vivo phage display
- PMID: 17784908
- PMCID: PMC2651023
- DOI: 10.1111/j.1365-2958.2007.05924.x
Borrelia burgdorferi adhesins identified using in vivo phage display
Abstract
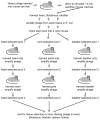
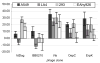
Borrelia burgdorferi, the agent of Lyme disease, disseminates from the site of deposition by Ixodes ticks to cause systemic infection. Dissemination occurs through the circulation and through tissue matrices, but the B. burgdorferi molecules that mediate interactions with the endothelium in vivo have not yet been identified. In vivo selection of filamentous phage expressing B. burgdorferi protein fragments on the phage surface identified several new candidate adhesins, and verified the activity of one adhesin that had been previously characterized in vitro. P66, a B. burgdorferi ligand for beta(3)-chain integrins, OspC, a protein that is essential for the establishment of infection in mammals, and Vls, a protein that undergoes antigenic variation in the mammal, were all selected for binding to the murine endothelium in vivo. Additional B. burgdorferi proteins for which no functions have been identified, including all four members of the OspF family and BmpD, were identified as candidate adhesins. The use of in vivo phage display is one approach to the identification of adhesins in pathogenic bacteria that are not easily grown in the laboratory, or for which genetic manipulations are not straightforward.
Figures

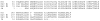
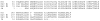


References
-
- Akins DR, Porcella SF, Popova TG, Shevchenko D, Baker SI, Li M, Norgard MV, Radolf JD. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Molecular microbiology. 1995;18:507–520. - PubMed
-
- Alitalo A, Meri T, Lankinen H, Seppala I, Lahdenne P, Hefty PS, Akins D, Meri S. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J Immunol. 2002;169:3847–3853. - PubMed
-
- Anguita J, Ramamoorthi N, Hovius JW, Das S, Thomas V, Persinski R, Conze D, Askenase PW, Rincon M, Kantor FS, Fikrig E. Salp15, an ixodes scapularis salivary protein, inhibits CD4+ T cell activation. Immunity. 2002;16:849–859. - PubMed
-
- Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. - PubMed
-
- Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

