Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere
- PMID: 17784941
- PMCID: PMC2375017
- DOI: 10.1186/gb-2007-8-9-r179
Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere
Abstract
Background: Mutualistic interactions less well known than those between rhizobia and legumes are commonly found between plants and bacteria, frequently pseudomonads, which colonize roots and adjacent soil areas (the rhizosphere).
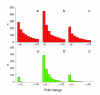
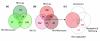
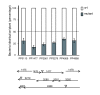
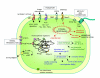
Results: A global analysis of Pseudomonas putida genes expressed during their interaction with maize roots revealed how a bacterial population adjusts its genetic program to this lifestyle. Differentially expressed genes were identified by comparing rhizosphere-colonizing populations with three distinct controls covering a variety of nutrients, growth phases and life styles (planktonic and sessile). Ninety rhizosphere up-regulated (rup) genes, which were induced relative to all three controls, were identified, whereas there was no repressed gene in common between the experiments. Genes involved in amino acid uptake and metabolism of aromatic compounds were preferentially expressed in the rhizosphere, which reflects the availability of particular nutrients in root exudates. The induction of efflux pumps and enzymes for glutathione metabolism indicates that adaptation to adverse conditions and stress (oxidative) response are crucial for bacterial life in this environment. The finding of a GGDEF/EAL domain response regulator among the induced genes suggests a role for the turnover of the secondary messenger c-diGMP in root colonization. Several mutants in rup genes showed reduced fitness in competitive root colonization.
Conclusion: Our results show the importance of two selective forces of different nature to colonize the rhizosphere: stress adaptation and availability of particular nutrients. We also identify new traits conferring bacterial survival in this niche and open a way to the characterization of specific signalling and regulatory processes governing the plant-Pseudomonas association.
Figures
References
-
- Molina LA, Ramos C, Duque E, Ronchel MC, García JM, Wyke L, Ramos JL. Survival of Pseudomonas putida KT2440 in soil and in the rhizosphere of plants under greenhouse and environmental conditions. Soil Biol Biochem. 2000;32:315–321. doi: 10.1016/S0038-0717(99)00156-X. - DOI
-
- Marilley L, Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. doi: 10.1016/S0929-1393(99)00028-1. - DOI
-
- Lugtenberg BJJ, Bloemberg GV. Life in the rhizosphere. In: Ramos JL, editor. Pseudomonas. Vol. 1. New York: Kluver; 2004. pp. 403–430.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

