Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae
- PMID: 17785441
- PMCID: PMC2169055
- DOI: 10.1128/MCB.01331-07
Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae
Abstract
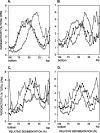
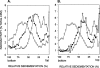
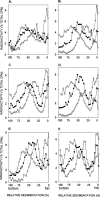
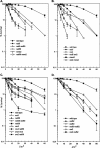
In Saccharomyces cerevisiae, replication through DNA lesions is promoted by Rad6-Rad18-dependent processes that include translesion synthesis by DNA polymerases eta and zeta and a Rad5-Mms2-Ubc13-controlled postreplicational repair (PRR) pathway which repairs the discontinuities in the newly synthesized DNA that form opposite from DNA lesions on the template strand. Here, we examine the contributions of the RAD51, RAD52, and RAD54 genes and of the RAD50 and XRS2 genes to the PRR of UV-damaged DNA. We find that deletions of the RAD51, RAD52, and RAD54 genes impair the efficiency of PRR and that almost all of the PRR is inhibited in the absence of both Rad5 and Rad52. We suggest a role for the Rad5 pathway when the lesion is located on the leading strand template and for the Rad52 pathway when the lesion is located on the lagging strand template. We surmise that both of these pathways operate in a nonrecombinational manner, Rad5 by mediating replication fork regression and template switching via its DNA helicase activity and Rad52 via a synthesis-dependent strand annealing mode. In addition, our results suggest a role for the Rad50 and Xrs2 proteins and thereby for the MRX complex in promoting PRR via both the Rad5 and Rad52 pathways.
Figures

Similar articles
-
Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Dec;27(23):8409-18. doi: 10.1128/MCB.01543-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923688 Free PMC article.
-
Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2002 Apr;22(7):2419-26. doi: 10.1128/MCB.22.7.2419-2426.2002. Mol Cell Biol. 2002. PMID: 11884624 Free PMC article.
-
Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae.Mol Cell Biol. 2006 Oct;26(20):7783-90. doi: 10.1128/MCB.01260-06. Epub 2006 Aug 14. Mol Cell Biol. 2006. PMID: 16908531 Free PMC article.
-
DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae.Mutat Res. 2001 Aug 9;486(3):167-84. doi: 10.1016/s0921-8777(01)00091-x. Mutat Res. 2001. PMID: 11459630 Review.
-
Error-free DNA-damage tolerance in Saccharomyces cerevisiae.Mutat Res Rev Mutat Res. 2015 Apr-Jun;764:43-50. doi: 10.1016/j.mrrev.2015.02.001. Epub 2015 Feb 16. Mutat Res Rev Mutat Res. 2015. PMID: 26041265 Review.
Cited by
-
Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae.Mol Cell Biol. 2007 Dec;27(23):8409-18. doi: 10.1128/MCB.01543-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923688 Free PMC article.
-
Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae.Genetics. 2010 Jan;184(1):65-77. doi: 10.1534/genetics.109.111039. Epub 2009 Nov 9. Genetics. 2010. PMID: 19901069 Free PMC article.
-
Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA.Mol Cell Biol. 2010 Feb;30(3):684-93. doi: 10.1128/MCB.00863-09. Epub 2009 Nov 30. Mol Cell Biol. 2010. PMID: 19948885 Free PMC article.
-
Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch.PLoS Genet. 2010 Nov 11;6(11):e1001205. doi: 10.1371/journal.pgen.1001205. PLoS Genet. 2010. PMID: 21085632 Free PMC article.
-
Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis.EMBO J. 2013 Mar 6;32(5):742-55. doi: 10.1038/emboj.2013.9. Epub 2013 Feb 8. EMBO J. 2013. PMID: 23395907 Free PMC article.
References
-
- Amitani, I., R. J. Baskin, and S. C. Kowalczykowski. 2006. Visualization of Rad52, a chromatin remodeling protein, translocating on single DNA molecules. Mol. Cell 23: 143-148. - PubMed
-
- Anderson, D. E., K. M. Trujillo, P. Sung, and H. P. Erickson. 2001. Structure of the Rad50-Mre11 DNA repair complex from Saccharomyces cerevisiae by electron microscopy. J. Biol. Chem. 276: 37027-37033. - PubMed
-
- Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8: 811-820. - PubMed
-
- Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272: 23360-23365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
