Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis
- PMID: 17785453
- PMCID: PMC2169052
- DOI: 10.1128/MCB.00002-07
Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis
Abstract
Mammalian DNA polymerase delta (Pol delta) is believed to replicate a large portion of the genome and to synthesize DNA in DNA repair and genetic recombination pathways. The effects of mutation in the polymerase domain of this essential enzyme are unknown. Here, we generated mice harboring an L604G or L604K substitution in highly conserved motif A in the polymerase active site of Pol delta. Homozygous Pold1(L604G/L604G) and Pold1(L604K/L604K) mice died in utero. However, heterozygous animals were viable and displayed no overall increase in disease incidence, indicative of efficient compensation for the defective mutant polymerase. The life spans of wild-type and heterozygous Pold1(+/L604G) mice did not differ, while that of Pold1(+/L604K) mice was reduced by 18%. Cultured embryonic fibroblasts from the heterozygous strains exhibited comparable increases in both spontaneous mutation rate and chromosome aberrations. We observed no significant increase in cancer incidence; however, Pold1(+/L604K) mice bearing histologically diagnosed tumors died at a younger median age than wild-type mice. Our results indicate that heterozygous mutation at L604 in the polymerase active site of DNA polymerase delta reduces life span, increases genomic instability, and accelerates tumorigenesis in an allele-specific manner, novel findings that have implications for human cancer.
Figures

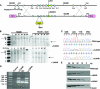
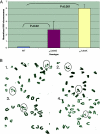

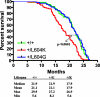


References
-
- Albertson, T. M., and B. D. Preston. 2006. DNA replication fidelity: proofreading in trans. Curr. Biol. 16: R209-R211. - PubMed
-
- Bebenek, K., C. M. Joyce, M. P. Fitzgerald, and T. A. Kunkel. 1990. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 265: 13878-13887. - PubMed
-
- Bland, M. 1995. An introduction to medical statistics, 2nd ed. Oxford University Press, Oxford, United Kingdom.
-
- Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276: 42462-42467. - PubMed
-
- Cullmann, G., R. Hindges, M. W. Berchtold, and U. Hubscher. 1993. Cloning of a mouse cDNA encoding DNA polymerase delta: refinement of the homology boxes. Gene 134: 191-200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
