Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads
- PMID: 17785517
- PMCID: PMC2064622
- DOI: 10.1083/jcb.200703106
Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads
Abstract
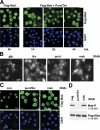
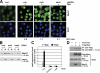
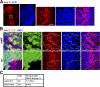
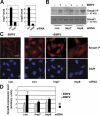
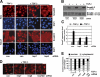
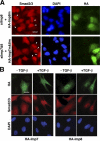
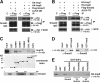
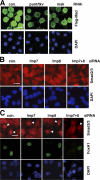
Nuclear translocation of Smad proteins is a critical step in signal transduction of transforming growth factor beta (TGF-beta) and bone morphogenetic proteins (BMPs). Using nuclear accumulation of the Drosophila Smad Mothers against Decapentaplegic (Mad) as the readout, we carried out a whole-genome RNAi screening in Drosophila cells. The screen identified moleskin (msk) as important for the nuclear import of phosphorylated Mad. Genetic evidence in the developing eye imaginal discs also demonstrates the critical functions of msk in regulating phospho-Mad. Moreover, knockdown of importin 7 and 8 (Imp7 and 8), the mammalian orthologues of Msk, markedly impaired nuclear accumulation of Smad1 in response to BMP2 and of Smad2/3 in response to TGF-beta. Biochemical studies further suggest that Smads are novel nuclear import substrates of Imp7 and 8. We have thus identified new evolutionarily conserved proteins that are important in the signal transduction of TGF-beta and BMP into the nucleus.
Figures
Similar articles
-
Specific nucleoporin requirement for Smad nuclear translocation.Mol Cell Biol. 2010 Aug;30(16):4022-34. doi: 10.1128/MCB.00124-10. Epub 2010 Jun 14. Mol Cell Biol. 2010. PMID: 20547758 Free PMC article.
-
Preferential utilization of Imp7/8 in nuclear import of Smads.J Biol Chem. 2008 Aug 15;283(33):22867-74. doi: 10.1074/jbc.M801320200. Epub 2008 Jun 2. J Biol Chem. 2008. PMID: 18519565 Free PMC article.
-
Identification of phosphatases for Smad in the BMP/DPP pathway.Genes Dev. 2006 Mar 15;20(6):648-53. doi: 10.1101/gad.1384706. Epub 2006 Mar 1. Genes Dev. 2006. PMID: 16510868 Free PMC article.
-
Nucleocytoplasmic shuttling of Smad proteins.Cell Res. 2009 Jan;19(1):36-46. doi: 10.1038/cr.2008.325. Cell Res. 2009. PMID: 19114992 Review.
-
Bone morphogenetic proteins.Growth Factors. 2004 Dec;22(4):233-41. doi: 10.1080/08977190412331279890. Growth Factors. 2004. PMID: 15621726 Review.
Cited by
-
Linderapyrone analogue LPD-01 as a cancer treatment agent by targeting importin7.J Nat Med. 2024 Mar;78(2):370-381. doi: 10.1007/s11418-023-01774-y. Epub 2024 Jan 24. J Nat Med. 2024. PMID: 38265612
-
Bi-allelic variants in IPO8 cause a connective tissue disorder associated with cardiovascular defects, skeletal abnormalities, and immune dysregulation.Am J Hum Genet. 2021 Jun 3;108(6):1126-1137. doi: 10.1016/j.ajhg.2021.04.020. Epub 2021 May 18. Am J Hum Genet. 2021. PMID: 34010604 Free PMC article.
-
Smad inhibition by the Ste20 kinase Misshapen.Proc Natl Acad Sci U S A. 2011 Jul 5;108(27):11127-32. doi: 10.1073/pnas.1104128108. Epub 2011 Jun 20. Proc Natl Acad Sci U S A. 2011. PMID: 21690388 Free PMC article.
-
Decoding the quantitative nature of TGF-beta/Smad signaling.Trends Cell Biol. 2008 Sep;18(9):430-42. doi: 10.1016/j.tcb.2008.06.006. Epub 2008 Aug 15. Trends Cell Biol. 2008. PMID: 18706811 Free PMC article. Review.
-
Genomic screening with RNAi: results and challenges.Annu Rev Biochem. 2010;79:37-64. doi: 10.1146/annurev-biochem-060408-092949. Annu Rev Biochem. 2010. PMID: 20367032 Free PMC article. Review.
References
-
- Adam, S.A., R. Sterne-Marr, and L. Gerace. 1992. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 219:97–110. - PubMed
-
- Armknecht, S., M. Boutros, A. Kiger, K. Nybakken, B. Mathey-Prevot, and N. Perrimon. 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392:55–73. - PubMed
-
- Batut, J., M. Howell, and C.S. Hill. 2007. Kinesin-mediated transport of smad2 is required for signaling in response to tgf-beta ligands. Dev. Cell. 12:261–274. - PubMed
-
- Chacko, B.M., B.Y. Qin, A. Tiwari, G. Shi, S. Lam, L.J. Hayward, M. De Caestecker, and K. Lin. 2004. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol. Cell. 15:813–823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

