Causes for the intriguing presence of tRNAs in phages
- PMID: 17785533
- PMCID: PMC1987346
- DOI: 10.1101/gr.6649807
Causes for the intriguing presence of tRNAs in phages
Abstract
Phages have highly compact genomes with sizes reflecting their capacity to exploit the host resources. Here, we investigate the reasons for tRNAs being the only translation-associated genes frequently found in phages. We were able to unravel the selective processes shaping the tRNA distribution in phages by analyzing their genomes and those of their hosts. We found ample evidence against tRNAs being selected to facilitate phage integration in the prokaryotic chromosomes. Conversely, there is a significant association between tRNA distribution and codon usage. We support this observation by introducing a master equation model, where tRNAs are randomly gained from their hosts and then lost either neutrally or according to a set of different selection mechanisms. Those tRNAs present in phages tend to correspond to codons that are simultaneously highly used by the phage genes, while rare in the host genome. Accordingly, we propose that a selective recruitment of tRNAs compensates for the compositional differences between the phage and the host genomes. To further understand the importance of these results in phage biology, we analyzed the differences between temperate and virulent phages. Virulent phages contain more tRNAs than temperate ones, higher codon usage biases, and more important compositional differences with respect to the host genome. These differences are thus in perfect agreement with the results of our master equation model and further suggest that tRNA acquisition may contribute to higher virulence. Thus, even though phages use most of the cell's translation machinery, they can complement it with their own genetic information to attain higher fitness. These results suggest that similar selection pressures may act upon other cellular essential genes that are being found in the recently uncovered large viruses.
Figures
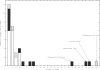
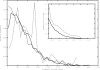
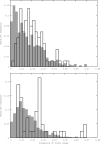
References
-
- Bailly-Bechet M., Danchin A., Iqbal M., Marsili M., Vergassola M., Danchin A., Iqbal M., Marsili M., Vergassola M., Iqbal M., Marsili M., Vergassola M., Marsili M., Vergassola M., Vergassola M. Codon usage domains over bacterial chromosomes. PLoS Comp. Biol. 2006;2:e37. doi: 10.1371/journal.pcbi.0020037. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
