Deletion of ultraconserved elements yields viable mice
- PMID: 17803355
- PMCID: PMC1964772
- DOI: 10.1371/journal.pbio.0050234
Deletion of ultraconserved elements yields viable mice
Abstract
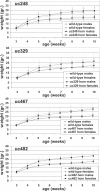
Ultraconserved elements have been suggested to retain extended perfect sequence identity between the human, mouse, and rat genomes due to essential functional properties. To investigate the necessities of these elements in vivo, we removed four noncoding ultraconserved elements (ranging in length from 222 to 731 base pairs) from the mouse genome. To maximize the likelihood of observing a phenotype, we chose to delete elements that function as enhancers in a mouse transgenic assay and that are near genes that exhibit marked phenotypes both when completely inactivated in the mouse and when their expression is altered due to other genomic modifications. Remarkably, all four resulting lines of mice lacking these ultraconserved elements were viable and fertile, and failed to reveal any critical abnormalities when assayed for a variety of phenotypes including growth, longevity, pathology, and metabolism. In addition, more targeted screens, informed by the abnormalities observed in mice in which genes in proximity to the investigated elements had been altered, also failed to reveal notable abnormalities. These results, while not inclusive of all the possible phenotypic impact of the deleted sequences, indicate that extreme sequence constraint does not necessarily reflect crucial functions required for viability.
Conflict of interest statement
Figures

Comment in
-
Are "ultraconserved" genetic elements really indispensable?PLoS Biol. 2007 Sep;5(9):e253. doi: 10.1371/journal.pbio.0050253. Epub 2007 Sep 4. PLoS Biol. 2007. PMID: 20076686 Free PMC article. No abstract available.
References
-
- Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. - PubMed
-
- Dermitzakis ET, Reymond A, Antonarakis SE. Conserved non-genic sequences—An unexpected feature of mammalian genomes. Nat Rev Genet. 2005;6:151–157. - PubMed
-
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, et al. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

