A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases
- PMID: 17803937
- PMCID: PMC2094128
- DOI: 10.1016/j.molcel.2007.06.028
A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases
Abstract
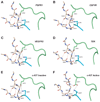
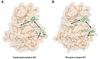
Activating mutations in the tyrosine kinase domain of receptor tyrosine kinases (RTKs) cause cancer and skeletal disorders. Comparison of the crystal structures of unphosphorylated and phosphorylated wild-type FGFR2 kinase domains with those of seven unphosphorylated pathogenic mutants reveals an autoinhibitory "molecular brake" mediated by a triad of residues in the kinase hinge region of all FGFRs. Structural analysis shows that many other RTKs, including PDGFRs, VEGFRs, KIT, CSF1R, FLT3, TEK, and TIE, are also subject to regulation by this brake. Pathogenic mutations activate FGFRs and other RTKs by disengaging the brake either directly or indirectly.
Figures





References
-
- Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. - PubMed
-
- Bellus GA, McIntosh I, Smith EA, Aylsworth AS, Kaitila I, Horton WA, Greenhaw GA, Hecht JT, Francomano CA. A recurrent mutation in the tyrosine kinase domain of fibroblast growth factor receptor 3 causes hypochondroplasia. Nat Genet. 1995;10:357–359. - PubMed
-
- Bellus GA, Spector EB, Speiser PW, Weaver CA, Garber AT, Bryke CR, Israel J, Rosengren SS, Webster MK, Donoghue DJ, Francomano CA. Distinct missense mutations of the FGFR3 lys650 codon modulate receptor kinase activation and the severity of the skeletal dysplasia phenotype. Am J Hum Genet. 2000;67:1411–1421. - PMC - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Calvert JT, Riney TJ, Kontos CD, Cha EH, Prieto VG, Shea CR, Berg JN, Nevin NC, Simpson SA, Pasyk KA, et al. Allelic and locus heterogeneity in inherited venous malformations. Hum Mol Genet. 1999;8:1279–1289. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

