Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
- PMID: 17804015
- PMCID: PMC2048488
- DOI: 10.1016/j.jmb.2007.07.062
Requirement of helix P2.2 and nucleotide G1 for positioning the cleavage site and cofactor of the glmS ribozyme
Abstract
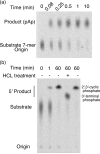
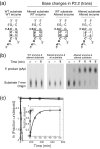
The glmS ribozyme is a catalytic RNA that self-cleaves at its 5'-end in the presence of glucosamine 6-phosphate (GlcN6P). We present structures of the glmS ribozyme from Thermoanaerobacter tengcongensis that are bound with the cofactor GlcN6P or the inhibitor glucose 6-phosphate (Glc6P) at 1.7 A and 2.2 A resolution, respectively. The two structures are indistinguishable in the conformations of the small molecules and of the RNA. GlcN6P binding becomes apparent crystallographically when the pH is raised to 8.5, where the ribozyme conformation is identical with that observed previously at pH 5.5. A key structural feature of this ribozyme is a short duplex (P2.2) that is formed between sequences just 3' of the cleavage site and within the core domain, and which introduces a pseudoknot into the active site. Mutagenesis indicates that P2.2 is required for activity in cis-acting and trans-acting forms of the ribozyme. P2.2 formation in a trans-acting ribozyme was exploited to demonstrate that N1 of the guanine at position 1 contributes to GlcN6P binding by interacting with the phosphate of the cofactor. At neutral pH, RNAs with adenine, 2-aminopurine, dimethyladenine or purine substitutions at position 1 cleave faster with glucosamine than with GlcN6P. This altered cofactor preference provides biochemical support for the orientation of the cofactor within the active site. Our results establish two features of the glmS ribozyme that are important for its activity: a sequence within the core domain that selects and positions the cleavage-site sequence, and a nucleobase at position 1 that helps position GlcN6P.
Figures


Similar articles
-
Protonation states of the key active site residues and structural dynamics of the glmS riboswitch as revealed by molecular dynamics.J Phys Chem B. 2010 Jul 8;114(26):8701-12. doi: 10.1021/jp9109699. J Phys Chem B. 2010. PMID: 20536206 Free PMC article.
-
Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme.Biochemistry. 2009 Apr 21;48(15):3239-46. doi: 10.1021/bi802069p. Biochemistry. 2009. PMID: 19228039 Free PMC article.
-
Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure.Nucleic Acids Res. 2006 Feb 7;34(3):968-75. doi: 10.1093/nar/gkj497. Print 2006. Nucleic Acids Res. 2006. PMID: 16464827 Free PMC article.
-
The glmS ribozyme: use of a small molecule coenzyme by a gene-regulatory RNA.Q Rev Biophys. 2010 Nov;43(4):423-47. doi: 10.1017/S0033583510000144. Epub 2010 Sep 8. Q Rev Biophys. 2010. PMID: 20822574 Free PMC article. Review.
-
Catalytic strategies of self-cleaving ribozymes.Acc Chem Res. 2008 Aug;41(8):1027-35. doi: 10.1021/ar800050c. Epub 2008 Jul 25. Acc Chem Res. 2008. PMID: 18652494 Review.
Cited by
-
Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules.Philos Trans R Soc Lond B Biol Sci. 2011 Oct 27;366(1580):2942-8. doi: 10.1098/rstb.2011.0131. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 21930586 Free PMC article.
-
Analysis of metal ion dependence in glmS ribozyme self-cleavage and coenzyme binding.Chembiochem. 2010 Dec 10;11(18):2567-71. doi: 10.1002/cbic.201000544. Chembiochem. 2010. PMID: 21108273 Free PMC article.
-
The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo.Nat Struct Mol Biol. 2011 Mar;18(3):359-63. doi: 10.1038/nsmb.1989. Epub 2011 Feb 13. Nat Struct Mol Biol. 2011. PMID: 21317896 Free PMC article.
-
Comprehensive sequence-to-function mapping of cofactor-dependent RNA catalysis in the glmS ribozyme.Nat Commun. 2020 Apr 3;11(1):1663. doi: 10.1038/s41467-020-15540-1. Nat Commun. 2020. PMID: 32245964 Free PMC article.
-
Protonation states of the key active site residues and structural dynamics of the glmS riboswitch as revealed by molecular dynamics.J Phys Chem B. 2010 Jul 8;114(26):8701-12. doi: 10.1021/jp9109699. J Phys Chem B. 2010. PMID: 20536206 Free PMC article.
References
-
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. - PubMed
-
- Klein DJ, Ferré-D'Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. - PubMed
-
- Edwards TE, Klein DJ, Ferré-D'Amaré AR. Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr. Op. Struct. Biol. 2007;17 in press. - PubMed
-
- Hampel KJ, Tinsley MM. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

