Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation
- PMID: 17804499
- PMCID: PMC2169032
- DOI: 10.1128/JVI.01123-07
Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation
Abstract
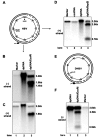
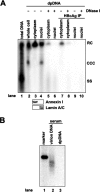
Covalently closed circular DNA (cccDNA) of hepatitis B virus (HBV) is formed by conversion of capsid-associated relaxed circular DNA (rcDNA) via unknown mechanisms and exists in the nucleus of the infected hepatocyte as a minichromosome that serves as the transcription template for viral RNAs. To study the molecular pathway of cccDNA formation and its regulation by viral and cellular factors, we have established a cell line that supports the replication of an envelope protein-deficient HBV genome in a tetracycline-inducible manner. Following induction of HBV replication, the cells accumulate higher levels of cccDNA as well as larger amounts of deproteinized rcDNA (DP-rcDNA) than cells that replicate wild-type HBV genomes. These results indicate that HBV envelope proteins negatively regulate cccDNA formation, and conversion of DP-rcDNA into cccDNA is a rate-limiting step of cccDNA formation in HepG2 cells. Detailed analyses reveal the following: (i) DP-rcDNA exists in both cytoplasm and nucleus; (ii) while nuclear DP-rcDNA is sensitive to DNase I digestion, a small fraction of cytoplasmic DP-rcDNA is DNase I resistant; (iii) both DNase I-sensitive and -resistant cytoplasmic DP-rcDNAs cosediment with capsids and can be immunoprecipitated with HBV core antibody; and (iv) a primer extension assay maps the 5' end of the minus strand of DP-rcDNA at the authentic end of virion rcDNA. Hence, our results favor a hypothesis that the removal of viral polymerase protein covalently linked to the 5' end of the minus-strand DNA occurs inside the capsid in the cytoplasm and most possibly via a reaction that cleaves the phosphodiester bond between the tyrosine of the polymerase and the 5' phosphoryl group of minus-strand DNA.
Figures




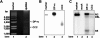



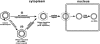
References
-
- Barthelmes, H. U., M. Habermeyer, M. O. Christensen, C. Mielke, H. Interthal, J. J. Pouliot, F. Boege, and D. Marko. 2004. TDP1 overexpression in human cells counteracts DNA damage mediated by topoisomerases I and II. J. Biol. Chem. 279:55618-55625. - PubMed
-
- Connelly, J. C., and D. R. Leach. 2004. Repair of DNA covalently linked to protein. Mol. Cell 13:307-316. - PubMed
-
- Dixon, K., and E. Kopras. 2004. Genetic alterations and DNA repair in human carcinogenesis. Semin. Cancer Biol. 14:441-448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

