High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants
- PMID: 17804504
- PMCID: PMC2169014
- DOI: 10.1128/JVI.01296-07
High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants
Abstract
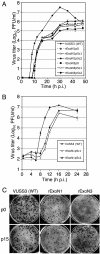
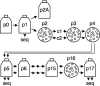
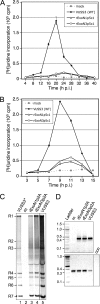
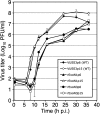
Replication fidelity of RNA virus genomes is constrained by the opposing necessities of generating sufficient diversity for adaptation and maintaining genetic stability, but it is unclear how the largest viral RNA genomes have evolved and are maintained under these constraints. A coronavirus (CoV) nonstructural protein, nsp14, contains conserved active-site motifs of cellular exonucleases, including DNA proofreading enzymes, and the severe acute respiratory syndrome CoV (SARS-CoV) nsp14 has 3'-to-5' exoribonuclease (ExoN) activity in vitro. Here, we show that nsp14 ExoN remarkably increases replication fidelity of the CoV murine hepatitis virus (MHV). Replacement of conserved MHV ExoN active-site residues with alanines resulted in viable mutant viruses with growth and RNA synthesis defects that during passage accumulated 15-fold more mutations than wild-type virus without changes in growth fitness. The estimated mutation rate for ExoN mutants was similar to that reported for other RNA viruses, whereas that of wild-type MHV was less than the established rates for RNA viruses in general, suggesting that CoVs with intact ExoN replicate with unusually high fidelity. Our results indicate that nsp14 ExoN plays a critical role in prevention or repair of nucleotide incorporation errors during genome replication. The established mutants are unique tools to test the hypothesis that high replication fidelity is required for the evolution and stability of large RNA genomes.
Figures



References
-
- Almazán, F., M. L. Dediego, C. Galán, D. Escors, E. Álvarez, J. Ortego, I. Sola, S. Zuñiga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80:10900-10906. - PMC - PubMed
-
- Bernad, A., L. Blanco, J. M. Lazaro, G. Martin, and M. Salas. 1989. A conserved 3′→5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell 59:219-228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

