Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes
- PMID: 17804629
- PMCID: PMC4916652
- DOI: 10.1523/JNEUROSCI.2750-07.2007
Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes
Abstract
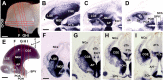
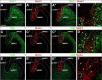
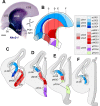
The mammalian telencephalon is considered the most complex of all biological structures. It comprises a large number of functionally and morphologically distinct types of neurons that coordinately control most aspects of cognition and behavior. The subpallium, for example, not only gives rise to multiple neuronal types that form the basal ganglia and parts of the amygdala and septum but also is the origin of an astonishing diversity of cortical interneurons. Despite our detailed knowledge on the molecular, morphological, and physiological properties of most of these neuronal populations, the mechanisms underlying their generation are still poorly understood. Here, we comprehensively analyzed the expression patterns of several transcription factors in the ventricular zone of the developing subpallium in the mouse to generate a detailed molecular map of the different progenitor domains present in this region. Our study demonstrates that the ventricular zone of the mouse subpallium contains at least 18 domains that are uniquely defined by the combinatorial expression of several transcription factors. Furthermore, the results of microtransplantation experiments in vivo corroborate that anatomically defined regions of the mouse subpallium, such as the medial ganglionic eminence, can be subdivided into functionally distinct domains.
Figures






References
-
- Bardet SM, Cobos I, Puelles E, Martinez-De-La-Torre M, Puelles L. Chicken lateral septal organ and other circumventricular organs form in a striatal subdomain abutting the molecular striatopallidal border. J Comp Neurol. 2006;499:745–767. - PubMed
-
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. - PMC - PubMed
-
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. - PubMed
-
- Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr Opin Neurobiol. 2003;13:50–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
