Centromere identity is specified by a single centromeric nucleosome in budding yeast
- PMID: 17804787
- PMCID: PMC1976213
- DOI: 10.1073/pnas.0706985104
Centromere identity is specified by a single centromeric nucleosome in budding yeast
Abstract
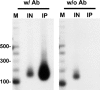
Chromosome segregation ensures that DNA is equally divided between daughter cells during each round of cell division. The centromere (CEN) is the specific locus on each chromosome that directs formation of the kinetochore, the multiprotein complex that interacts with the spindle microtubules to promote proper chromosomal alignment and segregation during mitosis. CENs are organized into a specialized chromatin structure due to the incorporation of an essential CEN-specific histone H3 variant (CenH3) in the centromeric nucleosomes of all eukaryotes. Consistent with its essential role at the CEN, the loss or up-regulation of CenH3 results in mitotic defects. Despite the requirement for CenH3 in CEN function, it is unclear how CenH3 nucleosomes structurally organize centromeric DNA to promote formation of the kinetochore. To address this issue, we developed a modified chromatin immunoprecipitation approach to analyze the number and position of CenH3 nucleosomes at the budding yeast CEN. Using this technique, we show that yeast CENs have a single CenH3 nucleosome positioned over the CEN-determining elements. Therefore, a single CenH3 nucleosome forms the minimal unit of centromeric chromatin necessary for kinetochore assembly and proper chromosome segregation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
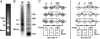
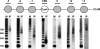
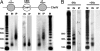
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

