Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism
- PMID: 17804808
- PMCID: PMC1976220
- DOI: 10.1073/pnas.0701989104
Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism
Abstract
Topoisomerase V is a type I topoisomerase without structural or sequence similarities to other topoisomerases. Although it belongs to the type I subfamily of topoisomerases, it is unrelated to either type IA or IB enzymes. We used real-time single-molecule micromechanical experiments to show that topoisomerase V relaxes DNA via events that release multiple DNA turns, employing a constrained swiveling mechanism similar to that for type IB enzymes. Relaxation is powered by the torque in the supercoiled DNA and is constrained by friction between the protein and the DNA. Although all type IB enzymes share a common structure and mechanism and type IA and type II enzymes show marked structural and functional similarities, topoisomerase V represents a different type of topoisomerase that relaxes DNA in a similar overall manner as type IB molecules but by using a completely different structural and mechanistic framework.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


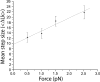
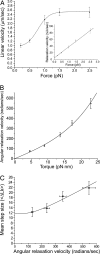
Similar articles
-
Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB.Nature. 2005 Mar 31;434(7033):671-4. doi: 10.1038/nature03395. Nature. 2005. PMID: 15800630
-
topIb, a phylogenetic hallmark gene of Thaumarchaeota encodes a functional eukaryote-like topoisomerase IB.Nucleic Acids Res. 2016 Apr 7;44(6):2795-805. doi: 10.1093/nar/gkw097. Epub 2016 Feb 22. Nucleic Acids Res. 2016. PMID: 26908651 Free PMC article.
-
DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote.Nature. 1993 Aug 19;364(6439):735-7. doi: 10.1038/364735a0. Nature. 1993. PMID: 8395022
-
[DNA supercoiling and topoisomerases in Escherichia coli].Rev Latinoam Microbiol. 1995 Jul-Sep;37(3):291-304. Rev Latinoam Microbiol. 1995. PMID: 8850348 Review. Spanish.
-
DNA topoisomerase: the key enzyme that regulates DNA super structure.Nagoya J Med Sci. 1998 May;61(1-2):11-26. Nagoya J Med Sci. 1998. PMID: 9664763 Review.
Cited by
-
A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity.Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):16125-30. doi: 10.1073/pnas.1206480109. Epub 2012 Sep 18. Proc Natl Acad Sci U S A. 2012. PMID: 22991469 Free PMC article.
-
Controlled rotation mechanism of DNA strand exchange by the Hin serine recombinase.Sci Rep. 2016 Apr 1;6:23697. doi: 10.1038/srep23697. Sci Rep. 2016. PMID: 27032966 Free PMC article.
-
A high-resolution magnetic tweezer for single-molecule measurements.Nucleic Acids Res. 2009 Nov;37(20):e136. doi: 10.1093/nar/gkp725. Epub 2009 Sep 3. Nucleic Acids Res. 2009. PMID: 19729511 Free PMC article.
-
Crossover-site sequence and DNA torsional stress control strand interchanges by the Bxb1 site-specific serine recombinase.Nucleic Acids Res. 2016 Oct 14;44(18):8921-8932. doi: 10.1093/nar/gkw724. Epub 2016 Aug 22. Nucleic Acids Res. 2016. PMID: 27550179 Free PMC article.
-
Mechanism of Type IA Topoisomerases.Molecules. 2020 Oct 17;25(20):4769. doi: 10.3390/molecules25204769. Molecules. 2020. PMID: 33080770 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

