Characterization of immune genes from the schistosome host snail Biomphalaria glabrata that encode peptidoglycan recognition proteins and gram-negative bacteria binding protein
- PMID: 17805526
- PMCID: PMC3632339
- DOI: 10.1007/s00251-007-0245-3
Characterization of immune genes from the schistosome host snail Biomphalaria glabrata that encode peptidoglycan recognition proteins and gram-negative bacteria binding protein
Abstract
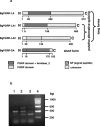
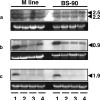
Peptidoglycan (PGN) recognition proteins (PGRPs) and gram-negative bacteria binding proteins (GNBPs) play an essential role in Toll/Imd signaling pathways in arthropods. The existence of homologous pathways involving PGRPs and GNBPs in other major invertebrate phyla such as the Mollusca remains unclear. In this paper, we report four full-length PGRP cDNAs and one full-length GNBP cDNA cloned from the snail Biomphalaria glabrata, the intermediate host of the human blood fluke Schistosoma mansoni, designated as BgPGRPs and BgGNBP, respectively. Three transcripts are generated from a long form PGRP gene (BgPGRP-LA) by alternative splicing and one from a short form PGRP gene (BgPGRP-SA). BgGNBP encodes a putative secreted protein. Northern blots demonstrated that expression of BgPGRP-SA and BgGNBP was down-regulated in B. glabrata at 6 h after exposure to three types of microbes. No significant changes in expression were observed in snails at 2 days post-exposure (dpe) to the trematodes Echinostoma paraensei or S. mansoni. However, up-regulation of BgPGRP-SA in M line snails at later time points of infection with E. paraensei (i.e., 12 and 17 dpe) was observed. Our study revealed that exposure to either microbes or trematodes did not alter the expression levels of BgPGRP-LAs, which were consistently low. This study provides new insights into the potential pathogen recognition capabilities of molluscs, indicates that further studies of the Toll/Imd pathways in this phylum are in order, and provides additional ways to judge the importance of this pathway in the evolution of internal defense across the animal phyla.
Figures





Similar articles
-
Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes).Mol Immunol. 2010 Jan;47(4):849-60. doi: 10.1016/j.molimm.2009.10.019. Epub 2009 Dec 3. Mol Immunol. 2010. PMID: 19962194 Free PMC article.
-
The effect of early infection with Echinostoma paraensei on the interaction of Schistosoma mansoni with Biomphalaria glabrata and Biomphalaria tenagophila.Mem Inst Oswaldo Cruz. 2010 Jul;105(4):499-503. doi: 10.1590/s0074-02762010000400026. Mem Inst Oswaldo Cruz. 2010. PMID: 20721499
-
Time series analysis of the transcriptional responses of Biomphalaria glabrata throughout the course of intramolluscan development of Schistosoma mansoni and Echinostoma paraensei.Int J Parasitol. 2010 Jun;40(7):819-31. doi: 10.1016/j.ijpara.2009.12.005. Epub 2010 Jan 18. Int J Parasitol. 2010. PMID: 20083115 Free PMC article.
-
Epigenetic modulation, stress and plasticity in susceptibility of the snail host, Biomphalaria glabrata, to Schistosoma mansoni infection.Int J Parasitol. 2016 Jun;46(7):389-94. doi: 10.1016/j.ijpara.2016.03.003. Epub 2016 Apr 4. Int J Parasitol. 2016. PMID: 27056272 Review.
-
[Immunity in parasite-vector snails].Med Sci (Paris). 2009 Apr;25(4):399-403. doi: 10.1051/medsci/2009254399. Med Sci (Paris). 2009. PMID: 19409193 Review. French.
Cited by
-
A haplotype-like, chromosome-level assembled and annotated genome of Biomphalaria glabrata, an important intermediate host of schistosomiasis and the best studied model of schistosomiasis vector snails.PLoS Negl Trop Dis. 2024 Feb 29;18(2):e0011983. doi: 10.1371/journal.pntd.0011983. eCollection 2024 Feb. PLoS Negl Trop Dis. 2024. PMID: 38421953 Free PMC article.
-
Molecular cloning and mRNA expression of the peptidoglycan recognition protein gene HcPGRP1 and its isoform HcPGRP1a from the freshwater mussel Hyriopsis cumingi.Genet Mol Biol. 2014 Sep;37(3):508-17. doi: 10.1590/s1415-47572014000400006. Genet Mol Biol. 2014. PMID: 25249773 Free PMC article.
-
H+ channels in embryonic Biomphalaria glabrata cell membranes: Putative roles in snail host-schistosome interactions.PLoS Negl Trop Dis. 2017 Mar 20;11(3):e0005467. doi: 10.1371/journal.pntd.0005467. eCollection 2017 Mar. PLoS Negl Trop Dis. 2017. PMID: 28319196 Free PMC article.
-
Identification and characterization of five transcription factors that are associated with evolutionarily conserved immune signaling pathways in the schistosome-transmitting snail Biomphalaria glabrata.Mol Immunol. 2011 Sep;48(15-16):1868-81. doi: 10.1016/j.molimm.2011.05.017. Epub 2011 Jun 21. Mol Immunol. 2011. PMID: 21696828 Free PMC article.
-
A genome sequence for Biomphalaria pfeifferi, the major vector snail for the human-infecting parasite Schistosoma mansoni.PLoS Negl Trop Dis. 2023 Mar 24;17(3):e0011208. doi: 10.1371/journal.pntd.0011208. eCollection 2023 Mar. PLoS Negl Trop Dis. 2023. PMID: 36961841 Free PMC article.
References
-
- Beschin A, Bilej M, Hanssens F, Raymakers J, Van Dyck E, Revets H, Brys L, Gomez J, De Baetselier P, Timmermans M. Identification and cloning of a glucan- and liopoplysaccharide-binding protein from Eisenia foetida earthworm involved in the activation of prophenoloxidase cascade. J Biol Chem. 1998;273:24948–24954. - PubMed
-
- Bilej M, De Baetselier P, Van Dijck E, Stijlemans B, Colige A, Beschin A. Distinct carbohydrate recognition domains of an invertebrate defense molecule recognize Gram-negative and Gram-positive bacteria. J Biol Chem. 2001;276:45840–45847. - PubMed
-
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. - PubMed
-
- Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

